4AMV
 
 | |
2J6H
 
 | |
3OOJ
 
 | |
2YJF
 
 | | Oligomeric assembly of actin bound to MRTF-A | | Descriptor: | ACTIN, ALPHA SKELETAL MUSCLE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Mouilleron, S, Langer, C.A, Guettler, S, McDonald, N.Q, Treisman, R. | | Deposit date: | 2011-05-19 | | Release date: | 2011-07-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of a pentavalent G-actin*MRTF-A complex reveals how G-actin controls nucleocytoplasmic shuttling of a transcriptional coactivator.
Sci Signal, 4, 2011
|
|
2YJE
 
 | | Oligomeric assembly of actin bound to MRTF-A | | Descriptor: | ACTIN, ALPHA SKELETAL MUSCLE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Mouilleron, S, Langer, C.A, Guettler, S, McDonald, N.Q, Treisman, R. | | Deposit date: | 2011-05-19 | | Release date: | 2011-07-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of a pentavalent G-actin*MRTF-A complex reveals how G-actin controls nucleocytoplasmic shuttling of a transcriptional coactivator.
Sci Signal, 4, 2011
|
|
2VF5
 
 | |
2V52
 
 | | Structure of MAL-RPEL2 complexed to G-actin | | Descriptor: | ACTIN, ALPHA SKELETAL MUSCLE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Mouilleron, S, Guettler, S, Langer, C.A, Treisman, R, McDonald, N.Q. | | Deposit date: | 2008-10-01 | | Release date: | 2008-11-25 | | Last modified: | 2018-02-28 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Molecular basis for G-actin binding to RPEL motifs from the serum response factor coactivator MAL.
EMBO J., 27, 2008
|
|
2V51
 
 | | Structure of MAL-RPEL1 complexed to actin | | Descriptor: | ACTIN, ALPHA SKELETAL MUSCLE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Mouilleron, S, Guettler, S, Langer, C.A, Treisman, R, McDonald, N.Q. | | Deposit date: | 2008-10-01 | | Release date: | 2008-11-25 | | Last modified: | 2018-02-28 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Molecular basis for G-actin binding to RPEL motifs from the serum response factor coactivator MAL.
EMBO J., 27, 2008
|
|
6YOO
 
 | | Structure of SAMM50 LIR bound to GABARAPL1 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Gamma-aminobutyric acid receptor-associated protein-like 1, ... | | Authors: | Mouilleron, S, Wenxin, Z, Johansen, T, Tooze, S, Abudu, Y.P, Lamark, T. | | Deposit date: | 2020-04-14 | | Release date: | 2021-04-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Structure of SAMM50 LIR bound to GABARAPL1
To Be Published
|
|
6YOP
 
 | | Structure of SAMM50 LIR bound to GABARAP | | Descriptor: | Sorting and assembly machinery component 50 homolog,Gamma-aminobutyric acid receptor-associated protein | | Authors: | Mouilleron, S, Wenxin, Z, Johansen, T, Tooze, S, Abudu, Y.P, Lamark, T. | | Deposit date: | 2020-04-14 | | Release date: | 2021-04-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structure of SAMM50 LIR bound to GABARAP
To Be Published
|
|
2VF4
 
 | | E. coli glucosamine-6-P synthase | | Descriptor: | GLUCOSAMINE--FRUCTOSE-6-PHOSPHATE AMINOTRANSFERASE | | Authors: | Mouilleron, S, Golinelli-Pimpaneau, B. | | Deposit date: | 2007-10-30 | | Release date: | 2008-03-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Ordering of C-Terminal Loop and Glutaminase Domains of Glucosamine-6-Phosphate Synthase Promotes Sugar Ring Opening and Formation of the Ammonia Channel.
J.Mol.Biol., 377, 2008
|
|
4B1Y
 
 | | Structure of the Phactr1 RPEL-3 bound to G-actin | | Descriptor: | ACTIN, ALPHA SKELETAL MUSCLE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Mouilleron, S, Wiezlak, M, O'Reilly, N, Treisman, R, McDonald, N.Q. | | Deposit date: | 2012-07-12 | | Release date: | 2013-07-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Structures of the Phactr1 RPEL domain and RPEL motif complexes with G-actin reveal the molecular basis for actin binding cooperativity.
Structure, 20, 2012
|
|
4B1Z
 
 | | Structure of the Phactr1 RPEL domain bound to G-actin | | Descriptor: | ACTIN, ALPHA SKELETAL MUSCLE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Mouilleron, S, Wiezlak, M, O'Reilly, N, Treisman, R, McDonald, N.Q. | | Deposit date: | 2012-07-12 | | Release date: | 2012-11-07 | | Last modified: | 2018-02-28 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structures of the Phactr1 RPEL domain and RPEL motif complexes with G-actin reveal the molecular basis for actin binding cooperativity.
Structure, 20, 2012
|
|
4B1W
 
 | | Structure of the Phactr1 RPEL-2 domain bound to actin | | Descriptor: | ACTIN, ALPHA SKELETAL MUSCLE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Mouilleron, S, Wiezlak, M, O'Reilly, N, Treisman, R, McDonald, N.Q. | | Deposit date: | 2012-07-12 | | Release date: | 2013-07-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of the Phactr1 RPEL domain and RPEL motif complexes with G-actin reveal the molecular basis for actin binding cooperativity.
Structure, 20, 2012
|
|
4B1U
 
 | | Structure of the Phactr1 RPEL domain and RPEL motif directed assemblies with G-actin reveal the molecular basis for actin binding cooperativity. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACTIN, ALPHA SKELETAL MUSCLE, ... | | Authors: | Mouilleron, S, Wiezlak, M, O'Reilly, N, Treisman, R, McDonald, N.Q. | | Deposit date: | 2012-07-12 | | Release date: | 2013-07-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of the Phactr1 RPEL domain and RPEL motif complexes with G-actin reveal the molecular basis for actin binding cooperativity.
Structure, 20, 2012
|
|
4B1X
 
 | | Structure of the Phactr1 RPEL-2 bound to G-actin | | Descriptor: | ACTIN, ALPHA SKELETAL MUSCLE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Mouilleron, S, Wiezlak, M, O'Reilly, N, Treisman, R, McDonald, N.Q. | | Deposit date: | 2012-07-12 | | Release date: | 2013-07-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of the Phactr1 RPEL domain and RPEL motif complexes with G-actin reveal the molecular basis for actin binding cooperativity.
Structure, 20, 2012
|
|
4B1V
 
 | | Structure of the Phactr1 RPEL-N domain bound to G-actin | | Descriptor: | 1,2-ETHANEDIOL, ACTIN, ALPHA SKELETAL MUSCLE, ... | | Authors: | Mouilleron, S, Wiezlak, M, O'Reilly, N, Treisman, R, McDonald, N.Q. | | Deposit date: | 2012-07-12 | | Release date: | 2012-11-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structures of the Phactr1 RPEL domain and RPEL motif complexes with G-actin reveal the molecular basis for actin binding cooperativity.
Structure, 20, 2012
|
|
5LXI
 
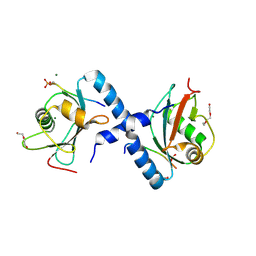 | | GABARAP-L1 ATG4B LIR Complex | | Descriptor: | 1,2-ETHANEDIOL, Cysteine protease ATG4B, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Mouilleron, S, Skytte Rasmussen, M, Kumar Shrestha, B, Wirth, M, Bowitz Larsen, K, Abudu Princely, Y, Sjottem, E, Tooze, S, Lamark, T, Johansen, T, Lee, R. | | Deposit date: | 2016-09-21 | | Release date: | 2017-02-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | ATG4B contains a C-terminal LIR motif important for binding and efficient cleavage of mammalian orthologs of yeast Atg8.
Autophagy, 13, 2017
|
|
5LXH
 
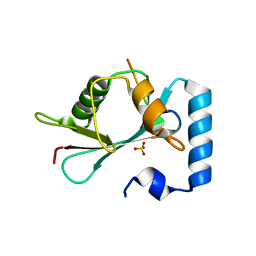 | | GABARAP-L1 ATG4B LIR Complex | | Descriptor: | Cysteine protease ATG4B, GLYCEROL, Gamma-aminobutyric acid receptor-associated protein-like 1, ... | | Authors: | Mouilleron, S, Skytte Rasmussen, M, Kumar Shrestha, B, Wirth, M, Bowitz Larsen, K, Abudu Princely, Y, Sjottem, E, Tooze, S, Lamark, T, Johansen, T, Lee, R. | | Deposit date: | 2016-09-21 | | Release date: | 2017-02-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | ATG4B contains a C-terminal LIR motif important for binding and efficient cleavage of mammalian orthologs of yeast Atg8.
Autophagy, 13, 2017
|
|
6GHM
 
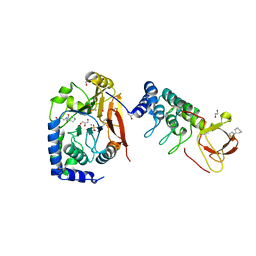 | | Structure of PP1 alpha phosphatase bound to ASPP2 | | Descriptor: | 1,2-ETHANEDIOL, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Apoptosis-stimulating of p53 protein 2, ... | | Authors: | Mouilleron, S, Bertran, T.M, Tapon, N, Zhou, Y. | | Deposit date: | 2018-05-08 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | ASPP proteins discriminate between PP1 catalytic subunits through their SH3 domain and the PP1 C-tail.
Nat Commun, 10, 2019
|
|
6ZEJ
 
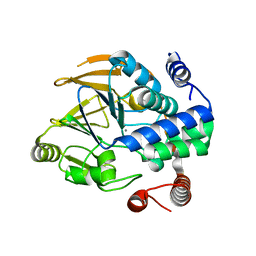 | | Structure of PP1-Phactr1 chimera [PP1(7-304) + linker (SGSGS) + Phactr1(526-580)] | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Mouilleron, S, Treisman, R, Fedoryshchak, R, Lee, R, Butler, A.M, Prechova, M. | | Deposit date: | 2020-06-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Molecular basis for substrate specificity of the Phactr1/PP1 phosphatase holoenzyme.
Elife, 9, 2020
|
|
6ZEI
 
 | | Structure of PP1-IRSp53 S455E chimera [PP1(7-304) + linker (G/S)x9 + IRSp53(449-465)] bound to Phactr1 (516-580) | | Descriptor: | GLYCEROL, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Mouilleron, S, Treisman, R, Fedoryshchak, R, Lee, R, Butler, A.M, Prechova, M. | | Deposit date: | 2020-06-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Molecular basis for substrate specificity of the Phactr1/PP1 phosphatase holoenzyme.
Elife, 9, 2020
|
|
6ZEH
 
 | | Structure of PP1-spectrin alpha II chimera [PP1(7-304) + linker (G/S)x9 + spectrin alpha II (1025-1039)] bound to Phactr1 (516-580) | | Descriptor: | MANGANESE (II) ION, PHOSPHATE ION, Phosphatase and actin regulator, ... | | Authors: | Mouilleron, S, Treisman, R, Fedoryshchak, R, Lee, R, Butler, A.M, Prechova, M. | | Deposit date: | 2020-06-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Molecular basis for substrate specificity of the Phactr1/PP1 phosphatase holoenzyme.
Elife, 9, 2020
|
|
6ZEE
 
 | | Structure of PP1(7-300) bound to Phactr1 (507-580) at pH8.4 | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15,18-HEXAOXAICOSANE, GLYCEROL, ... | | Authors: | Mouilleron, S, Treisman, R, Fedoryshchak, R, Lee, R, Butler, A.M, Prechova, M. | | Deposit date: | 2020-06-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular basis for substrate specificity of the Phactr1/PP1 phosphatase holoenzyme.
Elife, 9, 2020
|
|
6ZEF
 
 | | Structure of PP1(7-300) bound to Phactr1 (516-580) at pH 5.25 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, MANGANESE (II) ION, ... | | Authors: | Mouilleron, S, Treisman, R, Fedoryshchak, R, Lee, R, Butler, A.M, Prechova, M. | | Deposit date: | 2020-06-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Molecular basis for substrate specificity of the Phactr1/PP1 phosphatase holoenzyme.
Elife, 9, 2020
|
|
