1C6W
 
 | | MAUROCALCIN FROM SCORPIO MAURUS | | Descriptor: | MAUROCALCIN | | Authors: | Mosbah, A, Kharrat, R, Fajloun, Z, Renisio, J.-G, Blanc, E, Sabatier, J.-M, Ayeb, M, Darbon, H. | | Deposit date: | 1999-12-23 | | Release date: | 2000-09-06 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | A new fold in the scorpion toxin family, associated with an activity on a ryanodine-sensitive calcium channel.
Proteins, 40, 2000
|
|
1EHX
 
 | | NMR SOLUTION STRUCTURE OF THE LAST UNKNOWN MODULE OF THE CELLULOSOMAL SCAFFOLDIN PROTEIN CIPC OF CLOSTRIDUM CELLULOLYTICUM | | Descriptor: | SCAFFOLDIN PROTEIN | | Authors: | Mosbah, A, Belaich, A, Bornet, O, Belaich, J.P, Henrissat, B, Darbon, H. | | Deposit date: | 2000-02-23 | | Release date: | 2000-11-17 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the module X2 1 of unknown function of the cellulosomal scaffolding protein CipC of Clostridium cellulolyticum.
J.Mol.Biol., 304, 2000
|
|
1K19
 
 | | NMR Solution Structure of the Chemosensory Protein CSP2 from Moth Mamestra brassicae | | Descriptor: | Chemosensory Protein CSP2 | | Authors: | Mosbah, A, Campanacci, V, Lartigue, A, Tegoni, M, Cambillau, C, Darbon, H. | | Deposit date: | 2001-09-24 | | Release date: | 2002-12-04 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a chemosensory protein from the moth Mamestra brassicae
BIOCHEM.J., 369, 2003
|
|
1Q01
 
 | | Lebetin peptides, a new class of potent aggregation inhibitors | | Descriptor: | lebetin 2 isoform alpha | | Authors: | Mosbah, A, Marrakchi, N, Ganzalez, M.J, Van Rietschoten, J, Giralt, E, El Ayeb, M, Rochat, H, Sabatier, J.M, Darbon, H, Mabrouk, K. | | Deposit date: | 2003-07-15 | | Release date: | 2005-05-03 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Lebetin peptides, a new class of potent aggregation inhibitors
To be Published
|
|
1JMN
 
 | | Solution Structure of the Viscotoxin A2 | | Descriptor: | viscotoxin A2 | | Authors: | Mosbah, A, Coulon, A, Bernard, C, Urech, K, Rouge, P, Darbon, H. | | Deposit date: | 2001-07-19 | | Release date: | 2003-06-24 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Viscotoxin A2
TO BE PUBLISHED
|
|
1PJV
 
 | | Cobatoxin 1 from Centruroides noxius Scorpion venom: Chemical Synthesis, 3-D Structure in Solution, Pharmacology and Docking on K+ channels | | Descriptor: | Cobatoxin 1 | | Authors: | Mosbah, A, Jouirou, B, Visan, V, Grissmer, S, El Ayeb, M, Rochat, H, De Waard, M, Mabrouk, K, Sabatier, J.M. | | Deposit date: | 2003-06-03 | | Release date: | 2004-03-09 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Cobatoxin 1 from Centruroides noxius scorpion venom: chemical synthesis, three-dimensional structure in solution, pharmacology and docking on K+ channels.
Biochem.J., 377, 2004
|
|
1Y2P
 
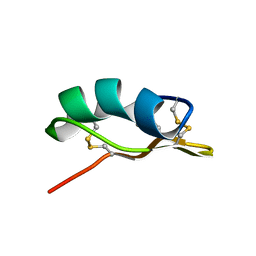 | |
1GMM
 
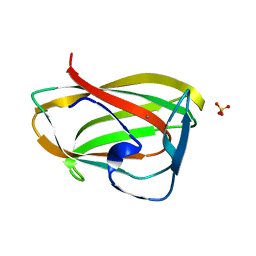 | | Carbohydrate binding module CBM6 from xylanase U Clostridium thermocellum | | Descriptor: | CALCIUM ION, CBM6, SODIUM ION, ... | | Authors: | Czjzek, M, Mosbah, A, Bolam, D, Allouch, J, Zamboni, V, Henrissat, B, Gilbert, H.J. | | Deposit date: | 2001-09-19 | | Release date: | 2001-11-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Location of the Ligand-Binding Site of Carbohydrate-Binding Modules that Have Evolved from a Common Sequence is not Conserved.
J.Biol.Chem., 276, 2001
|
|
1JMP
 
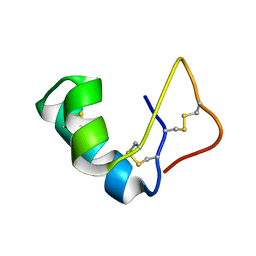 | | Solution Structure of the Viscotoxin B | | Descriptor: | viscotoxin B | | Authors: | Coulon, A, Mosbah, A, Bernard, C, Rouge, P, Urech, K, Darbon, H. | | Deposit date: | 2001-07-19 | | Release date: | 2003-11-11 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Comparative membrane interaction study of viscotoxins A3, A2 and B from mistletoe (Viscum album) and connections with their structures
Biochem.J., 374, 2003
|
|
1I26
 
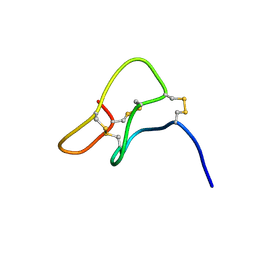 | | SOLUTION STRUCTURE OF PTU-1, A TOXIN FROM THE ASSASSIN BUGS PEIRATES TURPIS THAT BLOCKS THE VOLTAGE SENSITIVE CALCIUM CHANNEL N-TYPE | | Descriptor: | PTU-1 | | Authors: | Bernard, C, Corzo, G, Mosbah, A, Nakajima, T, Darbon, H. | | Deposit date: | 2001-02-06 | | Release date: | 2001-11-21 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Ptu1, a toxin from the assassin bug Peirates turpis that blocks the voltage-sensitive calcium channel N-type.
Biochemistry, 40, 2001
|
|
1WZ5
 
 | | Solution structure of Pi1-3p | | Descriptor: | Potassium channel blocking toxin 1 | | Authors: | Ferrat, G. | | Deposit date: | 2005-02-24 | | Release date: | 2005-04-19 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | The impact of the fourth disulfide bridge in scorpion toxins of the alpha-KTx6 subfamily
Proteins, 61, 2005
|
|
1KX9
 
 | | ANTENNAL CHEMOSENSORY PROTEIN A6 FROM THE MOTH MAMESTRA BRASSICAE | | Descriptor: | ACETATE ION, CHEMOSENSORY PROTEIN A6 | | Authors: | Lartigue, A, Campanacci, V, Roussel, A, Larsson, A.M, Jones, T.A, Tegoni, M, Cambillau, C. | | Deposit date: | 2002-01-31 | | Release date: | 2002-12-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | X-ray structure and ligand binding study of a moth chemosensory protein
J.Biol.Chem., 277, 2002
|
|
1KX8
 
 | | Antennal Chemosensory Protein A6 from Mamestra brassicae, tetragonal form | | Descriptor: | CHEMOSENSORY PROTEIN A6 | | Authors: | Lartigue, A, Campanacci, V, Roussel, A, Larsson, A.M, Jones, T.A, Tegoni, M, Cambillau, C. | | Deposit date: | 2002-01-31 | | Release date: | 2002-12-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-Ray Structure and Ligand Binding Study of a Chemosensory Protein
J.Biol.Chem., 277, 2002
|
|
