1NYF
 
 | |
1NYG
 
 | |
6XD4
 
 | | CDC-like protein | | Descriptor: | ACETATE ION, Hemolysin, SODIUM ION | | Authors: | Morton, C.J, Parker, M.W, Lawrence, S.L, Johnstone, B.A, Tweten, R.K. | | Deposit date: | 2020-06-09 | | Release date: | 2021-04-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Key Motif in the Cholesterol-Dependent Cytolysins Reveals a Large Family of Related Proteins.
Mbio, 11, 2020
|
|
3VDD
 
 | | Structure of HRV2 capsid complexed with antiviral compound BTA798 | | Descriptor: | 3-ethoxy-6-{2-[1-(6-methylpyridazin-3-yl)piperidin-4-yl]ethoxy}-1,2-benzoxazole, Protein VP1, Protein VP2, ... | | Authors: | Morton, C.J, Feil, S.C, Parker, M.W. | | Deposit date: | 2012-01-05 | | Release date: | 2012-09-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | An Orally Available 3-Ethoxybenzisoxazole Capsid Binder with Clinical Activity against Human Rhinovirus.
ACS Med Chem Lett, 3, 2012
|
|
7LXT
 
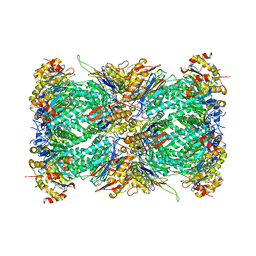 | | Structure of Plasmodium falciparum 20S proteasome with bound bortezomib | | Descriptor: | 20S proteasome alpha-1 subunit, 20S proteasome alpha-2 subunit, 20S proteasome alpha-3 subunit, ... | | Authors: | Morton, C.J, Metcalfe, R.D, Liu, B, Xie, S.C, Hanssen, E, Leis, A.P, Tilley, L, Griffin, M.D.W. | | Deposit date: | 2021-03-05 | | Release date: | 2021-09-22 | | Last modified: | 2022-04-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Design of proteasome inhibitors with oral efficacy in vivo against Plasmodium falciparum and selectivity over the human proteasome.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5IMT
 
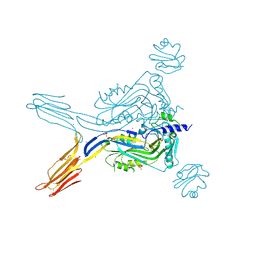 | | Toxin receptor complex | | Descriptor: | CD59 glycoprotein, COPPER (II) ION, Intermedilysin, ... | | Authors: | Morton, C.J, Lawrence, S.L, Feil, S.C, Parker, M.W. | | Deposit date: | 2016-03-06 | | Release date: | 2016-08-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7001 Å) | | Cite: | Structural Basis for Receptor Recognition by the Human CD59-Responsive Cholesterol-Dependent Cytolysins.
Structure, 24, 2016
|
|
6NAL
 
 | |
3CWL
 
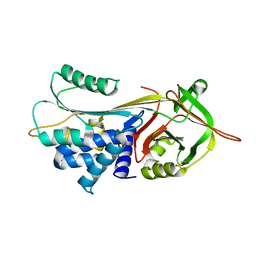 | | Crystal structure of alpha-1-antitrypsin, crystal form B | | Descriptor: | Alpha-1-antitrypsin, CHLORIDE ION | | Authors: | Morton, C.J, Hansen, G, Feil, S.C, Adams, J.J, Parker, M.W. | | Deposit date: | 2008-04-22 | | Release date: | 2008-09-23 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Preventing serpin aggregation: The molecular mechanism of citrate action upon antitrypsin unfolding.
Protein Sci., 17, 2008
|
|
3CWM
 
 | | Crystal structure of alpha-1-antitrypsin complexed with citrate | | Descriptor: | Alpha-1-antitrypsin, CITRIC ACID | | Authors: | Morton, C.J, Hansen, G, Feil, S.C, Adams, J.J, Parker, M.W. | | Deposit date: | 2008-04-22 | | Release date: | 2008-09-23 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Preventing serpin aggregation: The molecular mechanism of citrate action upon antitrypsin unfolding.
Protein Sci., 17, 2008
|
|
2QUG
 
 | | Crystal structure of alpha-1-antitrypsin, crystal form A | | Descriptor: | Alpha-1-antitrypsin | | Authors: | Hansen, G, Morton, C.J, Pearce, M.C, Feil, S.C, Adams, J.J, Parker, M.W, Bottomley, S.P. | | Deposit date: | 2007-08-05 | | Release date: | 2008-08-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Preventing serpin aggregation: The molecular mechanism of citrate action upon antitrypsin unfolding.
Protein Sci., 17, 2008
|
|
1A0N
 
 | | NMR STUDY OF THE SH3 DOMAIN FROM FYN PROTO-ONCOGENE TYROSINE KINASE COMPLEXED WITH THE SYNTHETIC PEPTIDE P2L CORRESPONDING TO RESIDUES 91-104 OF THE P85 SUBUNIT OF PI3-KINASE, FAMILY OF 25 STRUCTURES | | Descriptor: | FYN, PRO-PRO-ARG-PRO-LEU-PRO-VAL-ALA-PRO-GLY-SER-SER-LYS-THR | | Authors: | Renzoni, D.A, Pugh, D.J.R, Siligardi, G, Das, P, Morton, C.J, Rossi, C, Waterfield, M.D, Campbell, I.D, Ladbury, J.E. | | Deposit date: | 1997-12-05 | | Release date: | 1998-02-25 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Structural and thermodynamic characterization of the interaction of the SH3 domain from Fyn with the proline-rich binding site on the p85 subunit of PI3-kinase.
Biochemistry, 35, 1996
|
|
1AZG
 
 | | NMR STUDY OF THE SH3 DOMAIN FROM FYN PROTO-ONCOGENE TYROSINE KINASE KINASE COMPLEXED WITH THE SYNTHETIC PEPTIDE P2L CORRESPONDING TO RESIDUES 91-104 OF THE P85 SUBUNIT OF PI3-KINASE, MINIMIZED AVERAGE (PROBMAP) STRUCTURE | | Descriptor: | FYN, PRO-PRO-ARG-PRO-LEU-PRO-VAL-ALA-PRO-GLY-SER-SER-LYS-THR | | Authors: | Renzoni, D.A, Pugh, D.J.R, Siligardi, G, Das, P, Morton, C.J, Rossi, C, Waterfield, M.D, Campbell, I.D, Ladbury, J.E. | | Deposit date: | 1997-11-18 | | Release date: | 1998-02-25 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Structural and thermodynamic characterization of the interaction of the SH3 domain from Fyn with the proline-rich binding site on the p85 subunit of PI3-kinase.
Biochemistry, 35, 1996
|
|
7LXU
 
 | | Structure of Plasmodium falciparum 20S proteasome with bound MPI-5 | | Descriptor: | 20S proteasome alpha-1 subunit, 20S proteasome alpha-2 subunit, 20S proteasome alpha-3 subunit, ... | | Authors: | Metcalfe, R.D, Morton, C.J, Xie, S.C, Liu, B, Hanssen, E, Leis, A.P, Tilley, L, Griffin, M.D.W. | | Deposit date: | 2021-03-05 | | Release date: | 2021-09-22 | | Last modified: | 2022-04-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Design of proteasome inhibitors with oral efficacy in vivo against Plasmodium falciparum and selectivity over the human proteasome.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7LXV
 
 | | Structure of human 20S proteasome with bound MPI-5 | | Descriptor: | N-[(1R)-2-([1,1'-biphenyl]-4-yl)-1-boronoethyl]-1-methyl-L-prolinamide, Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, ... | | Authors: | Metcalfe, R.D, Morton, C.J, Liu, B, Xie, S.C, Hanssen, E, Leis, A.P, Tilley, L, Griffin, M.D.W. | | Deposit date: | 2021-03-05 | | Release date: | 2021-09-22 | | Last modified: | 2022-04-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Design of proteasome inhibitors with oral efficacy in vivo against Plasmodium falciparum and selectivity over the human proteasome.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5IMW
 
 | | Trapped Toxin | | Descriptor: | Intermedilysin | | Authors: | Lawrence, S.L, Feil, S.C, Morton, C.J, Parker, M.W. | | Deposit date: | 2016-03-07 | | Release date: | 2016-08-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structural Basis for Receptor Recognition by the Human CD59-Responsive Cholesterol-Dependent Cytolysins.
Structure, 24, 2016
|
|
5IMY
 
 | | Trapped Toxin | | Descriptor: | CD59 glycoprotein, Vaginolysin | | Authors: | Lawrence, S.L, Morton, C.J, Parker, M.W. | | Deposit date: | 2016-03-07 | | Release date: | 2016-08-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis for Receptor Recognition by the Human CD59-Responsive Cholesterol-Dependent Cytolysins.
Structure, 24, 2016
|
|
5TLQ
 
 | | Model structure of the oxidized PaDsbA1 and 3-[(2-methylbenzyl)sulfanyl]-4H-1,2,4-triazol-4-amine complex | | Descriptor: | 3-[(2-methylbenzyl)sulfanyl]-4H-1,2,4-triazol-4-amine, Thiol:disulfide interchange protein DsbA | | Authors: | Mohanty, B, Rimmer, K.A, McMahon, R.M, Headey, S.J, Vazirani, M, Shouldice, S.R, Coincon, M, Tay, S, Morton, C.J, Simpson, J.S, Martin, J.L, Scanlon, M.S. | | Deposit date: | 2016-10-11 | | Release date: | 2017-04-12 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Fragment library screening identifies hits that bind to the non-catalytic surface of Pseudomonas aeruginosa DsbA1.
PLoS ONE, 12, 2017
|
|
8TC8
 
 | | Human asparaginyl-tRNA synthetase bound to adenosine 5'-sulfamate | | Descriptor: | 5'-O-[N-(L-ASPARAGINYL)SULFAMOYL]ADENOSINE, Asparagine--tRNA ligase, cytoplasmic, ... | | Authors: | Dogovski, C, Metcalfe, R.D, Xie, S.C, Morton, C.J, Tilley, L, Griffin, M.D.W. | | Deposit date: | 2023-06-30 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Reaction hijacking inhibition of Plasmodium falciparum asparagine tRNA synthetase.
Nat Commun, 15, 2024
|
|
8TC7
 
 | | Human asparaginyl-tRNA synthetase, apo form | | Descriptor: | Asparagine--tRNA ligase, cytoplasmic, CHLORIDE ION, ... | | Authors: | Dogovski, C, Metcalfe, R.D, Xie, S.C, Morton, C.J, Tilley, L, Griffin, M.D.W. | | Deposit date: | 2023-06-30 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Reaction hijacking inhibition of Plasmodium falciparum asparagine tRNA synthetase.
Nat Commun, 15, 2024
|
|
8TC9
 
 | | Human asparaginyl-tRNA synthetase bound to OSM-S-106 | | Descriptor: | Asparagine--tRNA ligase, cytoplasmic, GLYCEROL, ... | | Authors: | Dogovski, C, Metcalfe, R.D, Xie, S.C, Morton, C.J, Tilley, L, Griffin, M.D.W. | | Deposit date: | 2023-06-30 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Reaction hijacking inhibition of Plasmodium falciparum asparagine tRNA synthetase.
Nat Commun, 15, 2024
|
|
4P8Q
 
 | | Crystal Structure of Human Insulin Regulated Aminopeptidase with Alanine in Active Site | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Leucyl-cystinyl aminopeptidase, ... | | Authors: | Hermans, S.J, Ascher, D.B, Hancock, N.C, Holien, J.K, Michell, B, Morton, C.J, Parker, M.W. | | Deposit date: | 2014-03-31 | | Release date: | 2014-12-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Crystal structure of human insulin-regulated aminopeptidase with specificity for cyclic peptides.
Protein Sci., 24, 2015
|
|
4PJ6
 
 | | Crystal Structure of Human Insulin Regulated Aminopeptidase with Lysine in Active Site | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LYSINE, ... | | Authors: | Hermans, S.J, Ascher, D.B, Hancock, N.C, Holien, J.K, Michell, B, Morton, C.J, Parker, M.W. | | Deposit date: | 2014-05-12 | | Release date: | 2014-12-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Crystal structure of human insulin-regulated aminopeptidase with specificity for cyclic peptides.
Protein Sci., 24, 2015
|
|
1AOT
 
 | | NMR STRUCTURE OF THE FYN SH2 DOMAIN COMPLEXED WITH A PHOSPHOTYROSYL PEPTIDE, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | FYN PROTEIN-TYROSINE KINASE, PHOSPHOTYROSYL PEPTIDE | | Authors: | Mulhern, T.D, Shaw, G.L, Morton, C.J, Day, A.J, Campbell, I.D. | | Deposit date: | 1997-07-10 | | Release date: | 1998-01-14 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | The SH2 domain from the tyrosine kinase Fyn in complex with a phosphotyrosyl peptide reveals insights into domain stability and binding specificity.
Structure, 5, 1997
|
|
1AOU
 
 | | NMR STRUCTURE OF THE FYN SH2 DOMAIN COMPLEXED WITH A PHOSPHOTYROSYL PEPTIDE, 22 STRUCTURES | | Descriptor: | FYN PROTEIN-TYROSINE KINASE, PHOSPHOTYROSYL PEPTIDE | | Authors: | Mulhern, T.D, Shaw, G.L, Morton, C.J, Day, A.J, Campbell, I.D. | | Deposit date: | 1997-07-10 | | Release date: | 1998-01-14 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | The SH2 domain from the tyrosine kinase Fyn in complex with a phosphotyrosyl peptide reveals insights into domain stability and binding specificity.
Structure, 5, 1997
|
|
6PRO
 
 | | MnSOD from Geobacillus stearothermophilus | | Descriptor: | MANGANESE (II) ION, Superoxide dismutase | | Authors: | Adams, J.J, Morton, C.J, Parker, M.W. | | Deposit date: | 2019-07-10 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.263 Å) | | Cite: | The Crystal Structure of the Manganese Superoxide Dismutase from Geobacillus stearothermophilus: Parker and Blake (1988) Revisited
Aust.J.Chem., 73, 2020
|
|
