1E8A
 
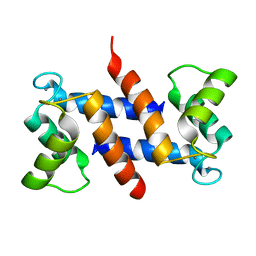 | | The three-dimensional structure of human S100A12 | | Descriptor: | CALCIUM ION, S100A12 | | Authors: | Moroz, O.V, Antson, A.A, Murshudov, G.N, Maitland, N.J, Dodson, G.G, Wilson, K.S, Skibshoj, I, Lukanidin, E.M, Bronstein, I.B. | | Deposit date: | 2000-09-18 | | Release date: | 2001-01-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Three-Dimensional Structure of Human S100A12
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1GQM
 
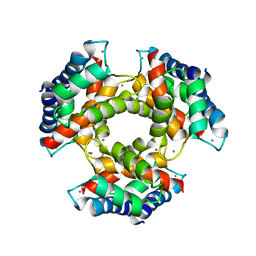 | | The structure of S100A12 in a hexameric form and its proposed role in receptor signalling | | Descriptor: | CALCIUM ION, CALGRANULIN C | | Authors: | Moroz, O.V, Antson, A.A, Dodson, E.G, Burrel, H.J, Grist, S.J, Lloyd, R.M, Maitland, N.J, Dodson, G.G, Wilson, K.S, Lukanidin, E, Bronstein, I.B. | | Deposit date: | 2001-11-26 | | Release date: | 2002-02-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Structure of S100A12 in a Hexameric Form and its Proposed Role in Receptor Signalling
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1ODB
 
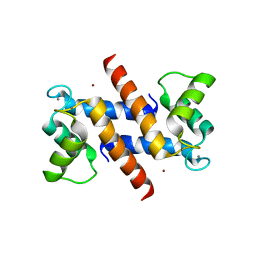 | | THE CRYSTAL STRUCTURE OF HUMAN S100A12 - COPPER COMPLEX | | Descriptor: | CALCIUM ION, CALGRANULIN C, COPPER (II) ION | | Authors: | Moroz, O.V, Antson, A.A, Grist, S.J, Maitland, N.J, Dodson, G.G, Wilson, K.S, Lukanidin, E.M, Bronstein, I.B. | | Deposit date: | 2003-02-15 | | Release date: | 2003-06-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structure of the Human S100A12-Copper Complex: Implications for Host-Parasite Defence
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1W2Y
 
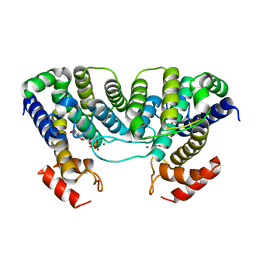 | | The crystal structure of a complex of Campylobacter jejuni dUTPase with substrate analogue dUpNHp | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-DIPHOSPHATE, DEOXYURIDINE 5'-TRIPHOSPHATE NUCLEOTIDE HYDROLASE, MAGNESIUM ION | | Authors: | Moroz, O.V, Harkiolaki, M, Galperin, M.Y, Vagin, A.A, Gonzalez-Pacanowska, D, Wilson, K.S. | | Deposit date: | 2004-07-09 | | Release date: | 2004-09-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Crystal Structure of a Complex of Campylobacter Jejuni Dutpase with Substrate Analogue Sheds Light on the Mechanism and Suggests the "Basic Module" for Dimeric D(C/U)Tpases
J.Mol.Biol., 342, 2004
|
|
4V20
 
 | | The 3-D structure of the cellobiohydrolase, Cel7A, from Aspergillus fumigatus, disaccharide complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, CELLOBIOHYDROLASE, ... | | Authors: | Moroz, O.V, Maranta, M, Shaghasi, T, Harris, P.V, Wilson, K.S, Davies, G.J. | | Deposit date: | 2014-10-05 | | Release date: | 2015-01-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Three-Dimensional Structure of the Cellobiohydrolase Cel7A from Aspergillus Fumigatus at 1.5 A Resolution
Acta Crystallogr.,Sect.F, 71, 2015
|
|
2WCE
 
 | | calcium-free (apo) S100A12 | | Descriptor: | PROTEIN S100-A12, SODIUM ION | | Authors: | Moroz, O.V, Blagova, E.V, Wilkinson, A.J, Wilson, K.S, Bronstein, I.B. | | Deposit date: | 2009-03-11 | | Release date: | 2009-06-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | The Crystal Structures of Human S100A12 in Apo Form and in Complex with Zinc: New Insights Into S100A12 Oligomerisation.
J.Mol.Biol., 391, 2009
|
|
2WCF
 
 | | calcium-free (apo) S100A12 | | Descriptor: | PROTEIN S100-A12, SODIUM ION | | Authors: | Moroz, O.V, Blagova, E.V, Wilkinson, A.J, Wilson, K.S, Bronstein, I.B. | | Deposit date: | 2009-03-11 | | Release date: | 2009-06-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | The Crystal Structures of Human S100A12 in Apo Form and in Complex with Zinc: New Insights Into S100A12 Oligomerisation.
J.Mol.Biol., 391, 2009
|
|
2WC8
 
 | | S100A12 complex with zinc in the absence of calcium | | Descriptor: | CITRIC ACID, PROTEIN S100-A12, SODIUM ION, ... | | Authors: | Moroz, O.V, Blagova, E.V, Wilkinson, A.J, Wilson, K.S, Bronstein, I.B. | | Deposit date: | 2009-03-10 | | Release date: | 2009-06-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | The Crystal Structures of Human S100A12 in Apo Form and in Complex with Zinc: New Insights Into S100A12 Oligomerisation.
J.Mol.Biol., 391, 2009
|
|
2WCB
 
 | | S100A12 complex with zinc in the absence of calcium | | Descriptor: | PROTEIN S100-A12, SODIUM ION, ZINC ION | | Authors: | Moroz, O.V, Blagova, E.V, Wilkinson, A.J, Wilson, K.S, Bronstein, I.B. | | Deposit date: | 2009-03-10 | | Release date: | 2009-06-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | The Crystal Structures of Human S100A12 in Apo Form and in Complex with Zinc: New Insights Into S100A12 Oligomerisation.
J.Mol.Biol., 391, 2009
|
|
4V1Z
 
 | | The 3-D structure of the cellobiohydrolase, Cel7A, from Aspergillus fumigatus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CELLOBIOHYDROLASE, ZINC ION | | Authors: | Moroz, O.V, Maranta, M, Shaghasi, T, Harris, P.V, Wilson, K.S, Davies, G.J. | | Deposit date: | 2014-10-04 | | Release date: | 2015-01-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | The Three-Dimensional Structure of the Cellobiohydrolase Cel7A from Aspergillus Fumigatus at 1.5 A Resolution
Acta Crystallogr.,Sect.F, 71, 2015
|
|
5FYP
 
 | | Calcium-dependent phosphoinositol-specific phospholipase C from a Gram-negative bacterium, Pseudomonas sp, apo form, crystal form 2 | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, PHOSPHOINOSITOL-SPECIFIC PHOSPHOLIPASE C, ... | | Authors: | Moroz, O.V, Blagova, E, Lebedev, A.A, Norgaard, A, Segura, D.R, Blicher, T.H, Wilson, K.S. | | Deposit date: | 2016-03-09 | | Release date: | 2017-01-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | The structure of a calcium-dependent phosphoinositide-specific phospholipase C from Pseudomonas sp. 62186, the first from a Gram-negative bacterium.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5FYR
 
 | | Calcium-dependent phosphoinositol-specific phospholipase C from a Gram-negative bacterium, Pseudomonas sp, apo form, myoinositol complex | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, CALCIUM ION, PHOSPHATE ION, ... | | Authors: | Moroz, O.V, Blagova, E, Lebedev, A.A, Norgaard, A, Segura, D.R, Blicher, T.H, Wilson, K.S. | | Deposit date: | 2016-03-09 | | Release date: | 2017-01-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The structure of a calcium-dependent phosphoinositide-specific phospholipase C from Pseudomonas sp. 62186, the first from a Gram-negative bacterium.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5FYO
 
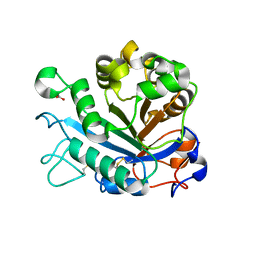 | | Calcium-dependent phosphoinositol-specific phospholipase C from a Gram-negative bacterium, Pseudomonas sp, apo form, crystal form 1 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, PHOSPHOINOSITOL-SPECIFIC PHOSPHOLIPASE C | | Authors: | Moroz, O.V, Blagova, E, Lebedev, A.A, Norgaard, A, Segura, D.R, Blicher, T.H, Wilson, K.S. | | Deposit date: | 2016-03-08 | | Release date: | 2017-01-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The structure of a calcium-dependent phosphoinositide-specific phospholipase C from Pseudomonas sp. 62186, the first from a Gram-negative bacterium.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
2Y5I
 
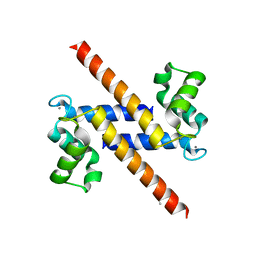 | | S100Z from zebrafish in complex with calcium | | Descriptor: | CALCIUM ION, ISOPROPYL ALCOHOL, S100 CALCIUM BINDING PROTEIN Z | | Authors: | Moroz, O.V, Bronstein, I.B, Wilson, K.S. | | Deposit date: | 2011-01-13 | | Release date: | 2011-07-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | The Crystal Structure of Zebrafish S100Z: Implications for Calcium-Promoted S100 Protein Oligomerisation.
J.Mol.Biol., 411, 2011
|
|
6ZMV
 
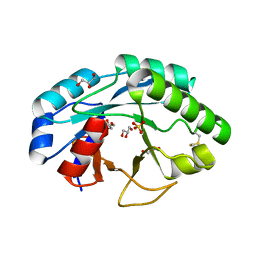 | | Structure of muramidase from Trichobolus zukalii | | Descriptor: | GLYCEROL, SULFATE ION, muramidase | | Authors: | Moroz, O.V, Blagova, E, Taylor, E, Turkenburg, J.P, Skov, L.K, Gippert, G.P, Schnorr, K.M, Ming, L, Ye, L, Klausen, M, Cohn, M.T, Schmidt, E.G.W, Nymand-Grarup, S, Davies, G.J, Wilson, K.S. | | Deposit date: | 2020-07-04 | | Release date: | 2021-07-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Fungal GH25 muramidases: New family members with applications in animal nutrition and a crystal structure at 0.78 angstrom resolution.
Plos One, 16, 2021
|
|
6ZM8
 
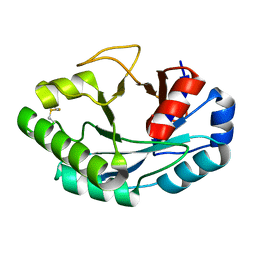 | | Structure of muramidase from Acremonium alcalophilum | | Descriptor: | muramidase | | Authors: | Moroz, O.V, Blagova, E, Taylor, E, Turkenburg, J.P, Skov, L.K, Gippert, G.P, Schnorr, K.M, Ming, L, Ye, L, Klausen, M, Cohn, M.T, Schmidt, E.G.W, Nymand-Grarup, S, Davies, G.J, Wilson, K.S. | | Deposit date: | 2020-07-01 | | Release date: | 2021-07-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (0.78 Å) | | Cite: | Fungal GH25 muramidases: New family members with applications in animal nutrition and a crystal structure at 0.78 angstrom resolution.
Plos One, 16, 2021
|
|
2C40
 
 | | CRYSTAL STRUCTURE OF INOSINE-URIDINE PREFERRING NUCLEOSIDE HYDROLASE FROM BACILLUS ANTHRACIS AT 2.2A RESOLUTION | | Descriptor: | CALCIUM ION, INOSINE-URIDINE PREFERRING NUCLEOSIDE HYDROLASE FAMILY PROTEIN, alpha-D-ribofuranose | | Authors: | Moroz, O.V, Blagova, E.V, Fogg, M.J, Levdikov, V.M, Brannigan, J.A, Wilkinson, A.J, Wilson, K.S. | | Deposit date: | 2005-10-13 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Inosine-Uridine Preferring Nucleoside Hydrolase from Bacillus Anthracis at 2.2A Resolution
To be Published
|
|
2BTU
 
 | | Crystal structure of Phosphoribosylformylglycinamidine cyclo-ligase from Bacillus Anthracis at 2.3A resolution. | | Descriptor: | PHOSPHORIBOSYL-AMINOIMIDAZOLE SYNTHETASE | | Authors: | Moroz, O.V, Blagova, E.V, Levdikov, V.M, Fogg, M.J, Lebedev, A.A, Brannigan, J.A, Wilkinson, A.J, Wilson, K.S. | | Deposit date: | 2005-06-07 | | Release date: | 2006-08-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Crystal Structure of Phosphoribosylformylglycinamidine Cyclo-Ligase from Bacillus Anthracis at 2.3A Resolution.
To be Published
|
|
6QPP
 
 | | Rhizomucor miehei lipase propeptide complex, native | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Lipase | | Authors: | Moroz, O.V, Blagova, E, Reiser, V, Saikia, R, Dalal, S, Jorgensen, C.I, Baunsgaard, L, Andersen, B, Svendsen, A, Wilson, K.S. | | Deposit date: | 2019-02-14 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Novel Inhibitory Function of theRhizomucor mieheiLipase Propeptide and Three-Dimensional Structures of Its Complexes with the Enzyme.
Acs Omega, 4, 2019
|
|
6QPR
 
 | | Rhizomucor miehei lipase propeptide complex, Ser95/Ile96 deletion mutant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Lipase | | Authors: | Moroz, O.V, Blagova, E, Reiser, V, Saikia, R, Dalal, S, Jorgensen, C.I, Baunsgaard, L, Andersen, B, Svendsen, A, Wilson, K.S. | | Deposit date: | 2019-02-14 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Novel Inhibitory Function of theRhizomucor mieheiLipase Propeptide and Three-Dimensional Structures of Its Complexes with the Enzyme.
Acs Omega, 4, 2019
|
|
7NIT
 
 | | X-ray structure of a multidomain BbgIII from Bifidobacterium bifidum | | Descriptor: | Beta-galactosidase, CALCIUM ION, GLYCEROL, ... | | Authors: | Moroz, O.V, Blagova, E, Lebedev, A.A, Sanchez Rodriguez, F, Rigden, D.J, Tams, J.W, Wilting, R, Vester, J.K, Longhin, E, Krogh, K.B.R, Pache, R.A, Davies, G.J, Wilson, K.S. | | Deposit date: | 2021-02-14 | | Release date: | 2021-12-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Multitasking in the gut: the X-ray structure of the multidomain BbgIII from Bifidobacterium bifidum offers possible explanations for its alternative functions.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
6F1J
 
 | | Structure of a Talaromyces pinophilus GH62 Arabinofuranosidase in complex with AraDNJ at 1.25A resolution | | Descriptor: | 1,4-DIDEOXY-1,4-IMINO-L-ARABINITOL, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Moroz, O.V, Sobala, L, Blagova, E, Coyle, T, Morkeberg Krogh, K.B.R, Wei, P, Stubbs, K, Wilson, K.S, Davies, G.J. | | Deposit date: | 2017-11-22 | | Release date: | 2018-08-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structure of a Talaromyces pinophilus GH62 arabinofuranosidase in complex with AraDNJ at 1.25 angstrom resolution.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
6FHN
 
 | | Structural dynamics and catalytic properties of a multi-modular xanthanase (Pt derivative) | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Moroz, O.V, Jensen, P.F, McDonald, S.P, McGregor, N, Blagova, E, Comamala, G, Segura, D.R, Anderson, L, Vasu, S.M, Rao, V.P, Giger, L, Monrad, R.N, Svendsen, A, Nielsen, J.E, Henrissat, B, Davies, G.J, Brumer, H, Rand, K, Wilson, K.S. | | Deposit date: | 2018-01-15 | | Release date: | 2018-08-29 | | Last modified: | 2019-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Dynamics and Catalytic Properties of a Multimodular Xanthanase
Acs Catalysis, 2018
|
|
6FHJ
 
 | | Structural dynamics and catalytic properties of a multi-modular xanthanase, native. | | Descriptor: | CALCIUM ION, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Moroz, O.V, Jensen, P.F, McDonald, S.P, McGregor, N, Blagova, E, Comamala, G, Segura, D.R, Anderson, L, Vasu, S.M, Rao, V.P, Giger, L, Monrad, R.N, Svendsen, A, Nielsen, J.E, Henrissat, B, Davies, G.J, Brumer, H, Rand, K, Wilson, K.S. | | Deposit date: | 2018-01-14 | | Release date: | 2018-08-29 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural Dynamics and Catalytic Properties of a Multimodular Xanthanase
Acs Catalysis, 2018
|
|
6G21
 
 | | Crystal structure of an esterase from Aspergillus oryzae | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(4-HYDROXY-3-METHOXYPHENYL)-2-PROPENOIC ACID, ... | | Authors: | Moroz, O.V, Blagova, E, Davies, G.J, Wilson, K.S. | | Deposit date: | 2018-03-22 | | Release date: | 2018-05-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of an esterase from Aspergillus oryzae
To Be Published
|
|
