5KAX
 
 | | The structure of CTR107 protein bound to RHODAMINE 6G | | Descriptor: | CTR107 protein, GLYCEROL, RHODAMINE 6G | | Authors: | Moreno, A, Wade, H. | | Deposit date: | 2016-06-02 | | Release date: | 2016-08-24 | | Last modified: | 2019-11-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Solution Binding and Structural Analyses Reveal Potential Multidrug Resistance Functions for SAV2435 and CTR107 and Other GyrI-like Proteins.
Biochemistry, 55, 2016
|
|
5KAU
 
 | | The structure of SAV2435 bound to RHODAMINE 6G | | Descriptor: | GLYCEROL, RHODAMINE 6G, SA2223 protein | | Authors: | Moreno, A, Wade, H. | | Deposit date: | 2016-06-02 | | Release date: | 2016-08-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Solution Binding and Structural Analyses Reveal Potential Multidrug Resistance Functions for SAV2435 and CTR107 and Other GyrI-like Proteins.
Biochemistry, 55, 2016
|
|
5KAW
 
 | |
5KAV
 
 | | The structure of SAV2435 | | Descriptor: | GLYCEROL, SA2223 protein | | Authors: | Moreno, A, Wade, H. | | Deposit date: | 2016-06-02 | | Release date: | 2016-08-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Solution Binding and Structural Analyses Reveal Potential Multidrug Resistance Functions for SAV2435 and CTR107 and Other GyrI-like Proteins.
Biochemistry, 55, 2016
|
|
5KAT
 
 | | The structure of SAV2435 bound to TETRAPHENYLPHOSPHONIUM | | Descriptor: | GLYCEROL, SA2223 protein, TETRAPHENYLPHOSPHONIUM | | Authors: | Moreno, A, Wade, H. | | Deposit date: | 2016-06-02 | | Release date: | 2016-08-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Solution Binding and Structural Analyses Reveal Potential Multidrug Resistance Functions for SAV2435 and CTR107 and Other GyrI-like Proteins.
Biochemistry, 55, 2016
|
|
5KCB
 
 | | The structure of SAV2435 bound to ethidium bromide | | Descriptor: | ETHIDIUM, SA2223 protein, SULFATE ION | | Authors: | Moreno, A, Wade, H. | | Deposit date: | 2016-06-06 | | Release date: | 2016-08-24 | | Last modified: | 2019-11-27 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Solution Binding and Structural Analyses Reveal Potential Multidrug Resistance Functions for SAV2435 and CTR107 and Other GyrI-like Proteins.
Biochemistry, 55, 2016
|
|
4UXM
 
 | | Crystal Structure of Struthiocalcin-1, a different crystal form. | | Descriptor: | (4-CARBOXYPHENYL)(CHLORO)MERCURY, STRUTHIOCALCIN-1 | | Authors: | Ruiz-Arellano, R.R, Moreno, A, Romero, A. | | Deposit date: | 2014-08-26 | | Release date: | 2015-04-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of Struthiocalcin-1, an Intramineral Protein from Struthio Camelus Eggshell, in Two Different Crystal Forms.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4UWW
 
 | | Crystallographic Structure of the Intramineral Protein Struthicalcin from Struthio camelus Eggshell | | Descriptor: | STRUTHIOCALCIN-1 | | Authors: | Ruiz, R.R, Moreno, A, Romero, A. | | Deposit date: | 2014-08-14 | | Release date: | 2015-04-08 | | Last modified: | 2015-04-22 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Crystal Structure of Struthiocalcin-1, an Intramineral Protein from Struthio Camelus Eggshell, in Two Different Crystal Forms.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
1GZ2
 
 | |
7Q1L
 
 | | Glycosilated Human Serum Apo-tranferrin | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Gavira, J.A, Moreno, A, Campos-Escamilla, C, Gonzalez-Ramirez, L.A, Siliqi, D. | | Deposit date: | 2021-10-20 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | X-ray Characterization of Conformational Changes of Human Apo- and Holo-Transferrin.
Int J Mol Sci, 22, 2021
|
|
2B4Z
 
 | | Crystal structure of cytochrome C from bovine heart at 1.5 A resolution. | | Descriptor: | Cytochrome c, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Mirkin, N, Jakoncic, J, Stojanoff, V, Moreno, A. | | Deposit date: | 2005-09-27 | | Release date: | 2005-10-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | High resolution X-ray crystallographic structure of bovine heart cytochrome c and its application to the design of an electron transfer biosensor.
Proteins, 70, 2008
|
|
4AFL
 
 | | The crystal structure of the ING4 dimerization domain reveals the functional organization of the ING family of chromatin binding proteins. | | Descriptor: | INHIBITOR OF GROWTH PROTEIN 4 | | Authors: | Culurgioni, S, Munoz, I.G, Moreno, A, Palacios, A, Villate, M, Palmero, I, Montoya, G, Blanco, F.J. | | Deposit date: | 2012-01-19 | | Release date: | 2012-02-22 | | Last modified: | 2012-07-11 | | Method: | X-RAY DIFFRACTION (2.275 Å) | | Cite: | Crystal Structure of Inhibitor of Growth 4 (Ing4) Dimerization Domain Reveals Functional Organization of Ing Family of Chromatin-Binding Proteins.
J.Biol.Chem., 287, 2012
|
|
2P3X
 
 | | Crystal structure of Grenache (Vitis vinifera) Polyphenol Oxidase | | Descriptor: | CU-O-CU LINKAGE, Polyphenol oxidase, chloroplast | | Authors: | Reyes Grajeda, J.P, Virador, V.M, Blanco-Labra, A, Mendiola-Olaya, E, Smith, G.M, Moreno, A, Whitaker, J.R. | | Deposit date: | 2007-03-09 | | Release date: | 2008-03-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Cloning, sequencing, purification, and crystal structure of Grenache (Vitis vinifera) polyphenol oxidase.
J.Agric.Food Chem., 58, 2010
|
|
5AIJ
 
 | | P. aeruginosa SdsA hexagonal polymorph | | Descriptor: | ALKYL SULFATASE, GLYCEROL, ZINC ION | | Authors: | De la Mora, E, Flores-Hernandez, E, Jakoncic, J, Stojanoff, V, Sanchez-Puig, N, Moreno, A. | | Deposit date: | 2015-02-13 | | Release date: | 2015-10-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Sdsa Polymorph Isolation and Improvement of Their Crystal Quality Using Nonconventional Crystallization Techniques
J.Appl.Crystallogr., 48, 2015
|
|
5A23
 
 | | SdsA sulfatase triclinic form | | Descriptor: | SDS HYDROLASE SDSA1, ZINC ION | | Authors: | De la Mora, E, Flores-Hernandez, E, Jakoncik, J, Stojanoff, V, Sanchez-Puig, N, Moreno, A. | | Deposit date: | 2015-05-11 | | Release date: | 2015-10-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Sdsa Polymorph Isolation and Improvement of Their Crystal Quality Using Nonconventional Crystallization Techniques
J.Appl.Crystallogr., 48, 2015
|
|
5AJL
 
 | | Sdsa sulfatase tetragonal | | Descriptor: | ALKYL SULFATASE, ZINC ION | | Authors: | De la Mora, E, Flores-Hernandez, E, Jakoncic, J, Stojanoff, V, Sanchez-Puig, N, Moreno, A. | | Deposit date: | 2015-02-25 | | Release date: | 2015-10-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Sdsa Polymorph Isolation and Improvement of Their Crystal Quality Using Nonconventional Crystallization Techniques
J.Appl.Crystallogr., 48, 2015
|
|
2M1R
 
 | | PHD domain of ING4 N214D mutant | | Descriptor: | Inhibitor of growth protein 4, ZINC ION | | Authors: | Blanco, F.J. | | Deposit date: | 2012-12-05 | | Release date: | 2012-12-26 | | Method: | SOLUTION NMR | | Cite: | Functional impact of cancer-associated mutations in the tumor suppressor protein ING4.
Carcinogenesis, 31, 2010
|
|
1CI1
 
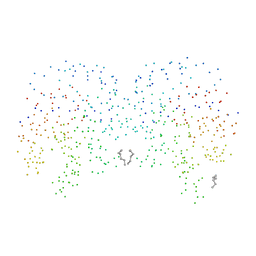 | | CRYSTAL STRUCTURE OF TRIOSEPHOSPHATE ISOMERASE FROM TRYPANOSOMA CRUZI IN HEXANE | | Descriptor: | HEXANE, PROTEIN (TRIOSEPHOSPHATE ISOMERASE) | | Authors: | Gao, X.-G, Maldondo, E, Perez-Montfort, R, De Gomez-Puyou, M.T, Gomez-Puyou, A, Rodriguez-Romero, A. | | Deposit date: | 1999-04-06 | | Release date: | 1999-09-01 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of triosephosphate isomerase from Trypanosoma cruzi in hexane.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1TCD
 
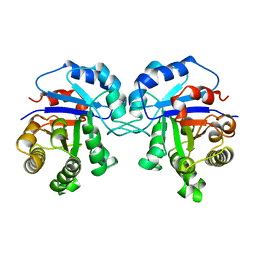 | | TRYPANOSOMA CRUZI TRIOSEPHOSPHATE ISOMERASE | | Descriptor: | TRIOSEPHOSPHATE ISOMERASE | | Authors: | Maldonado, E, Soriano-Garcia, M, Cabrera, N, Garza-Ramos, G, Tuena De Gomez-Puyou, M, Gomez-Puyou, A, Perez-Montfort, R. | | Deposit date: | 1998-01-29 | | Release date: | 1999-01-13 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Differences in the intersubunit contacts in triosephosphate isomerase from two closely related pathogenic trypanosomes.
J.Mol.Biol., 283, 1998
|
|
8GB5
 
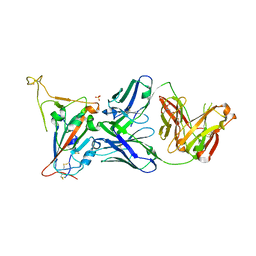 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing antibody 25F9 | | Descriptor: | 25F9 Heavy chain, 25F9 Light chain, BICINE, ... | | Authors: | Yuan, M, Zhu, X, Wilson, I.A. | | Deposit date: | 2023-02-24 | | Release date: | 2023-05-24 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Broadly neutralizing antibodies against sarbecoviruses generated by immunization of macaques with an AS03-adjuvanted COVID-19 vaccine.
Sci Transl Med, 15, 2023
|
|
8GB7
 
 | |
8GB6
 
 | |
8GB8
 
 | |
5EPM
 
 | |
5D1Z
 
 | | IsdB NEAT1 bound by clone D4-10 | | Descriptor: | D4-10 Heavy Chain, D4-10 Light Chain, Iron-regulated surface determinant protein B, ... | | Authors: | Deng, X. | | Deposit date: | 2015-08-04 | | Release date: | 2016-09-14 | | Last modified: | 2016-12-07 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | Germline-encoded neutralization of a Staphylococcus aureus virulence factor by the human antibody repertoire.
Nat Commun, 7, 2016
|
|
