1PG7
 
 | | Murine 6A6 Fab in complex with humanized anti-Tissue Factor D3H44 Fab | | Descriptor: | humanized antibody D3H44, murine antibody 6A6 Fab fragment | | Authors: | Eigenbrot, C, Meng, Y.G, Krishnamurthy, R, Lipari, M.T, Presta, L, Devaux, B, Wong, T, Moran, P, Bullens, S, Kirchhofer, D. | | Deposit date: | 2003-05-27 | | Release date: | 2003-08-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insight into how an anti-idiotypic antibody against D3H44 (anti-tissue factor antibody) restores normal coagulation.
J.Mol.Biol., 331, 2003
|
|
1SI5
 
 | | Protease-like domain from 2-chain hepatocyte growth factor | | Descriptor: | hepatocyte growth factor | | Authors: | Kirchhofer, D, Yao, X, Peek, M, Eigenbrot, C, Lipari, M.T, Billeci, K.L, Maun, H.R, Moran, P, Santell, L, Lazarus, R.A. | | Deposit date: | 2004-02-27 | | Release date: | 2004-12-28 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structural and functional basis of the serine protease-like hepatocyte growth factor beta-chain in Met binding and signaling
J.Biol.Chem., 279, 2004
|
|
7L6K
 
 | | ApoL1 N-terminal domain | | Descriptor: | Apolipoprotein L1 | | Authors: | Holliday, M.J, Ultsch, M, Moran, P, Fairbrother, W.J, Kirchhofer, D. | | Deposit date: | 2020-12-23 | | Release date: | 2021-08-04 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structures of the ApoL1 and ApoL2 N-terminal domains reveal a non-classical four-helix bundle motif.
Commun Biol, 4, 2021
|
|
4NMX
 
 | |
5VLK
 
 | |
5VLA
 
 | | Short PCSK9 delta-P' complex with Fusion2 peptide | | Descriptor: | CALCIUM ION, Proprotein convertase subtilisin/kexin type 9, THR-VAL-PHE-THR-SER-TRP-GLU-GLU-TYR-LEU-ASP-TRP-VAL-MET-PRO-TRP-ASN-LEU-VAL-ARG-ILE-GLY-LEU-LEU | | Authors: | Eigenbrot, C, Ultsch, M. | | Deposit date: | 2017-04-25 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of a cryptic peptide-binding site on PCSK9 and design of antagonists.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5VLP
 
 | | PCSK9 complex with LDLR antagonist peptide and Fab7G7 | | Descriptor: | Fab7G7 heavy chain, Fab7G7 light chain, LDLR antagonist peptide, ... | | Authors: | Eigenbrot, C, Ultsch, M. | | Deposit date: | 2017-04-25 | | Release date: | 2017-08-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery of a cryptic peptide-binding site on PCSK9 and design of antagonists.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5VLH
 
 | | Short PCSK9 delta-P' complex with peptide Pep1 | | Descriptor: | ACE-THR-VAL-PHE-THR-SER-TRP-GLU-GLU-TYR-LEU-ASP-TRP-VAL-NH2, CALCIUM ION, CYS-ARG-LEU-PRO-TRP-ASN-LEU-GLN-ARG-ILE-GLY-LEU-PRO-CYS, ... | | Authors: | Eigenbrot, C, Ultsch, M. | | Deposit date: | 2017-04-25 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Discovery of a cryptic peptide-binding site on PCSK9 and design of antagonists.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5VLL
 
 | | Short PCSK9 delta-P' complex with peptide Pep3 | | Descriptor: | ACE-THR-VAL-PHE-THR-SER-TRP-GLU-GLU-TYR-LEU-ASP-TRP-VAL-NH2, CALCIUM ION, CYS-PHE-ILE-PRO-TRP-ASN-LEU-GLN-ARG-ILE-GLY-LEU-LEU-CYS, ... | | Authors: | Eigenbrot, C, Ultsch, M. | | Deposit date: | 2017-04-25 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Discovery of a cryptic peptide-binding site on PCSK9 and design of antagonists.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5VL7
 
 | | PCSK9 complex with Fab33 | | Descriptor: | Fab33 heavy chain, Fab33 light chain, Proprotein convertase subtilisin/kexin type 9 | | Authors: | Eigenbrot, C, Shia, S. | | Deposit date: | 2017-04-25 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Discovery of a cryptic peptide-binding site on PCSK9 and design of antagonists.
Nat. Struct. Mol. Biol., 24, 2017
|
|
3P8X
 
 | | Synthesis, Structure, and Biological Activity of des-Side Chain Analogues of 1alpha,25-Dihydroxyvitamin D3 with Substituents at C-18 | | Descriptor: | (1R,3S,5Z)-5-{(2E)-2-[(3aR,7aS)-7a-(7-hydroxy-7-methyloctyl)octahydro-4H-inden-4-ylidene]ethylidene}-4-methylidenecyclohexane-1,3-diol, SULFATE ION, Vitamin D3 receptor | | Authors: | Rochel, N, Sato, Y, Moras, D. | | Deposit date: | 2010-10-15 | | Release date: | 2011-08-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Synthesis, Structure, and Biological Activity of des-Side Chain Analogues of 1 ,25-Dihydroxyvitamin D3 with Substituents at C-18
Chemmedchem, 6, 2011
|
|
1JPS
 
 | | Crystal structure of tissue factor in complex with humanized Fab D3h44 | | Descriptor: | immunoglobulin Fab D3H44, heavy chain, light chain, ... | | Authors: | Faelber, K, Kirchhofer, D, Presta, L, Kelley, R.F, Muller, Y.A. | | Deposit date: | 2001-08-03 | | Release date: | 2002-02-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The 1.85 A resolution crystal structures of tissue factor in complex with humanized Fab D3h44 and of free humanized Fab D3h44: revisiting the solvation of antigen combining sites.
J.Mol.Biol., 313, 2001
|
|
1JPT
 
 | | Crystal Structure of Fab D3H44 | | Descriptor: | immunoglobulin Fab D3H44, heavy chain, immunoglobulin Fab D3h44, ... | | Authors: | Faelber, K, Kirchhofer, D, Presta, L, Kelley, R.F, Muller, Y.A. | | Deposit date: | 2001-08-03 | | Release date: | 2002-02-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The 1.85 A resolution crystal structures of tissue factor in complex with humanized Fab D3h44 and of free humanized Fab D3h44: revisiting the solvation of antigen combining sites.
J.Mol.Biol., 313, 2001
|
|
3TJO
 
 | |
3TKC
 
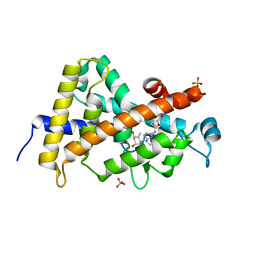 | | Design, Synthesis, Evaluation and Structure of Vitamin D Analogues with Furan Side Chains | | Descriptor: | (1S,3R,5Z,7E,14beta,17alpha,20S)-20-[5-(1-hydroxy-1-methylethyl)furan-2-yl]-9,10-secopregna-5,7,10-triene-1,3-diol, SULFATE ION, Vitamin D3 receptor | | Authors: | Huet, T, Moras, D, Rochel, N. | | Deposit date: | 2011-08-26 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Design, synthesis, evaluation, and structure of vitamin D analogues with furan side chains.
Chemistry, 18, 2012
|
|
3TJN
 
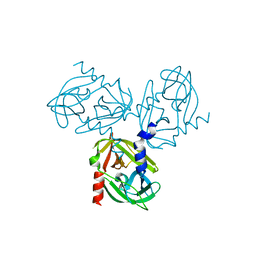 | | HtrA1 catalytic domain, apo form | | Descriptor: | Serine protease HTRA1 | | Authors: | Eigenbrot, C, Ultsch, M. | | Deposit date: | 2011-08-24 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and Functional Analysis of HtrA1 and Its Subdomains.
Structure, 20, 2012
|
|
3TJQ
 
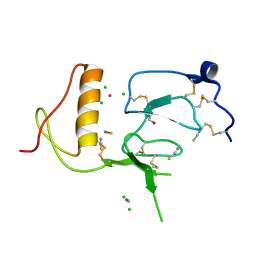 | | N-domain of HtrA1 | | Descriptor: | CHLORIDE ION, GLYCEROL, PLATINUM (II) ION, ... | | Authors: | Eigenbrot, C, Ultsch, M. | | Deposit date: | 2011-08-24 | | Release date: | 2012-05-16 | | Last modified: | 2012-06-27 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structural and Functional Analysis of HtrA1 and Its Subdomains.
Structure, 20, 2012
|
|
7LFA
 
 | | Fab 3B6 bound to ApoL1 NTD | | Descriptor: | Apolipoprotein L1, CHLORIDE ION, Fab 3B6 heavy chain, ... | | Authors: | Ultsch, M, Kirchhofer, D. | | Deposit date: | 2021-01-15 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.857 Å) | | Cite: | Structures of the ApoL1 and ApoL2 N-terminal domains reveal a non-classical four-helix bundle motif.
Commun Biol, 4, 2021
|
|
7LFB
 
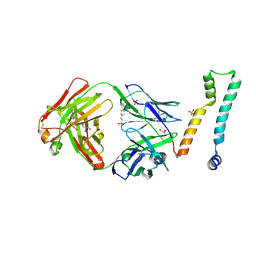 | | Fab 7D6 bound to ApoL1 NTD | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Apolipoprotein L1, Fab 7D6 heavy chain, ... | | Authors: | Ultsch, M, Kirchhofer, D. | | Deposit date: | 2021-01-16 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.913 Å) | | Cite: | Structures of the ApoL1 and ApoL2 N-terminal domains reveal a non-classical four-helix bundle motif.
Commun Biol, 4, 2021
|
|
7LF8
 
 | | Fab 6D12 bound to ApoL2 NTD | | Descriptor: | Apolipoprotein L2, Fab 6D12 heavy chain, Fab 6D12 light chain, ... | | Authors: | Ultsch, M, Kirchhofer, D. | | Deposit date: | 2021-01-15 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structures of the ApoL1 and ApoL2 N-terminal domains reveal a non-classical four-helix bundle motif.
Commun Biol, 4, 2021
|
|
7LFD
 
 | | Fab 7D6 bound to ApoL1 BH3 like peptide | | Descriptor: | AMMONIUM ION, Apolipoprotein L1 BH3 like peptide, CITRATE ANION, ... | | Authors: | Ultsch, M, Kirchhofer, D. | | Deposit date: | 2021-01-16 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.157 Å) | | Cite: | Structures of the ApoL1 and ApoL2 N-terminal domains reveal a non-classical four-helix bundle motif.
Commun Biol, 4, 2021
|
|
7LF7
 
 | | Fab 6D12 bound to ApoL1 NTD | | Descriptor: | Apolipoprotein L1, ETHANOL, Fab 6D12 heavy chain, ... | | Authors: | Ultsch, M, Kirchhofer, D. | | Deposit date: | 2021-01-15 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.026 Å) | | Cite: | Structures of the ApoL1 and ApoL2 N-terminal domains reveal a non-classical four-helix bundle motif.
Commun Biol, 4, 2021
|
|
1K6Q
 
 | | Crystal structure of antibody Fab fragment D3 | | Descriptor: | immunoglobulin Fab D3, heavy chain, light chain | | Authors: | Faelber, K, Kelley, R.F, Kirchhofer, D, Muller, Y.A. | | Deposit date: | 2001-10-17 | | Release date: | 2002-04-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of murine Fab D3 at 2.4 A resolution in comparison with the humanised Fab D3h44 (1.85A) provides structural insight into the humanisation process of the D3 anti-tissue factor antibody
To be Published
|
|
1SHY
 
 | |
