8GJV
 
 | |
1RZG
 
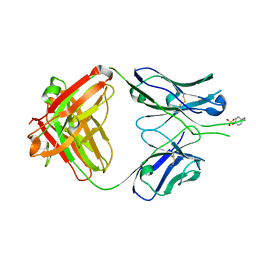 | | Crystal structure of Human anti-HIV-1 GP120 reactive antibody 412d | | Descriptor: | ASPARTIC ACID, CYSTEINE, Fab 412d heavy chain, ... | | Authors: | Huang, C.C, Venturi, M, Majeed, S, Moore, M.J, Phogat, S, Zhang, M.-Y, Dimitrov, D.S, Hendrickson, W.A, Robinson, J, Sodroski, J, Wyatt, R, Choe, H, Farzan, M, Kwong, P.D. | | Deposit date: | 2003-12-24 | | Release date: | 2004-02-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of tyrosine sulfation and VH-gene usage in antibodies that recognize the HIV type 1 coreceptor-binding site on gp120
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1RZ7
 
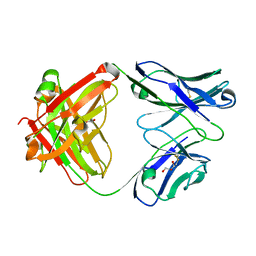 | | CRYSTAL STRUCTURE OF HUMAN ANTI-HIV-1 GP120-REACTIVE ANTIBODY 48D | | Descriptor: | Fab 48d heavy chain, Fab 48d light chain, GLYCEROL | | Authors: | Huang, C.C, Venturi, M, Majeed, S, Moore, M.J, Phogat, S, Zhang, M.-Y, Dimitrov, D.S, Hendrickson, W.A, Robinson, J, Sodroski, J, Wyatt, R, Choe, H, Farzan, M, Kwong, P.D. | | Deposit date: | 2003-12-24 | | Release date: | 2004-02-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of tyrosine sulfation and VH-gene usage in antibodies that recognize the HIV type 1 coreceptor-binding site on gp120
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1RZI
 
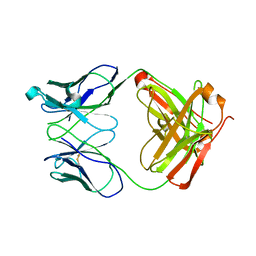 | | Crystal structure of human anti-HIV-1 gp120-reactive antibody 47e fab | | Descriptor: | Fab 47e heavy chain, Fab 47e light chain | | Authors: | Huang, C.C, Venturi, M, Majeed, S, Moore, M.J, Phogat, S, Zhang, M.-Y, Dimitrov, D.S, Hendrickson, W.A, Robinson, J, Sodroski, J, Wyatt, R, Choe, H, Farzan, M, Kwong, P.D. | | Deposit date: | 2003-12-24 | | Release date: | 2004-02-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of tyrosine sulfation and VH-gene usage in antibodies that recognize the HIV type 1 coreceptor-binding site on gp120
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1RZF
 
 | | Crystal structure of Human anti-HIV-1 GP120-reactive antibody E51 | | Descriptor: | Fab E51 heavy chain, Fab E51 light chain, GLYCEROL, ... | | Authors: | Huang, C.C, Venturi, M, Majeed, S, Moore, M.J, Phogat, S, Zhang, M.-Y, Dimitrov, D.S, Hendrickson, W.A, Robinson, J, Sodroski, J, Wyatt, R, Choe, H, Farzan, M, Kwong, P.D. | | Deposit date: | 2003-12-24 | | Release date: | 2004-02-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of tyrosine sulfation and VH-gene usage in antibodies that recognize the HIV type 1 coreceptor-binding site on gp120
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1RZ8
 
 | | CRYSTAL STRUCTURE OF HUMAN ANTI-HIV-1 GP120-REACTIVE ANTIBODY 17B | | Descriptor: | Fab 17b heavy chain, Fab 17b light chain | | Authors: | Huang, C.C, Venturi, M, Majeed, S, Moore, M.J, Phogat, S, Zhang, M.-Y, Dimitrov, D.S, Hendrickson, W.A, Robinson, J, Sodroski, J, Wyatt, R, Choe, H, Farzan, M, Kwong, P.D. | | Deposit date: | 2003-12-24 | | Release date: | 2004-02-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of tyrosine sulfation and VH-gene usage in antibodies that recognize the HIV type 1 coreceptor-binding site on gp120
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1RZK
 
 | | HIV-1 YU2 GP120 ENVELOPE GLYCOPROTEIN COMPLEXED WITH CD4 AND INDUCED NEUTRALIZING ANTIBODY 17B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ANTIBODY 17B, HEAVY CHAIN, ... | | Authors: | Huang, C.C, Venturi, M, Majeed, S, Moore, M.J, Phogat, S, Zhang, M.-Y, Dimitrov, D.S, Hendrickson, W.A, Robinson, J, Sodroski, J, Wyatt, R, Choe, H, Farzan, M, Kwong, P.D. | | Deposit date: | 2003-12-24 | | Release date: | 2004-02-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of tyrosine sulfation and VH-gene usage in antibodies that recognize the HIV type 1 coreceptor-binding site on gp120
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1RZJ
 
 | | HIV-1 HXBC2 GP120 ENVELOPE GLYCOPROTEIN COMPLEXED WITH CD4 AND INDUCED NEUTRALIZING ANTIBODY 17B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ANTIBODY 17B, HEAVY CHAIN, ... | | Authors: | Huang, C.C, Venturi, M, Majeed, S, Moore, M.J, Phogat, S, Zhang, M.-Y, Dimitrov, D.S, Hendrickson, W.A, Robinson, J, Sodroski, J, Wyatt, R, Choe, H, Farzan, M, Kwong, P.D. | | Deposit date: | 2003-12-24 | | Release date: | 2004-02-03 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of tyrosine sulfation and VH-gene usage in antibodies that recognize the HIV type 1 coreceptor-binding site on gp120
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
8U4N
 
 | | Structure of Apo CXCR4/Gi complex | | Descriptor: | C-X-C chemokine receptor type 4, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Saotome, K, McGoldrick, L.L, Franklin, M.C. | | Deposit date: | 2023-09-11 | | Release date: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Structural insights into CXCR4 modulation and oligomerization
Biorxiv, 2024
|
|
8U4S
 
 | |
8U4T
 
 | |
8U4P
 
 | |
8U4Q
 
 | |
8U4R
 
 | |
8U4O
 
 | |
8ES9
 
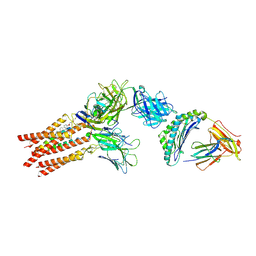 | | CryoEM structure of PN45428 TCR-CD3 in complex with HLA-A2 MAGEA4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, ... | | Authors: | Saotome, K, Franklin, M.C. | | Deposit date: | 2022-10-13 | | Release date: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Structural analysis of cancer-relevant TCR-CD3 and peptide-MHC complexes by cryoEM
Nat Commun, 14, 2023
|
|
8ESB
 
 | |
8ES7
 
 | | CryoEM structure of PN45545 TCR-CD3 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Saotome, K, Franklin, M.C. | | Deposit date: | 2022-10-13 | | Release date: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Structural analysis of cancer-relevant TCR-CD3 and peptide-MHC complexes by cryoEM
Nat Commun, 14, 2023
|
|
8ES8
 
 | |
8ESA
 
 | |
