6RXH
 
 | | In-flow serial synchrotron crystallography using a 3D-printed microfluidic device (3D-MiXD): Aspartate alpha-decarboxylase | | Descriptor: | Aspartate 1-decarboxylase, UNKNOWN ATOM OR ION | | Authors: | Monteiro, D.C.F, von Stetten, D, Pearson, A.R, Trebbin, M. | | Deposit date: | 2019-06-08 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 3D-MiXD: 3D-printed X-ray-compatible microfluidic devices for rapid, low-consumption serial synchrotron crystallography data collection in flow.
Iucrj, 7, 2020
|
|
6RXI
 
 | | In-flow serial synchrotron crystallography using a 3D-printed microfluidic device (3D-MiXD): Lysozyme | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Lysozyme C, ... | | Authors: | Monteiro, D.C.F, von Stetten, D, Pearson, A.R, Trebbin, M. | | Deposit date: | 2019-06-08 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 3D-MiXD: 3D-printed X-ray-compatible microfluidic devices for rapid, low-consumption serial synchrotron crystallography data collection in flow.
Iucrj, 7, 2020
|
|
8FUH
 
 | |
8FVV
 
 | |
8FXD
 
 | |
6H79
 
 | |
4CRY
 
 | | Direct visualisation of strain-induced protein post-translational modification | | Descriptor: | ACETYL COENZYME *A, ASPARTATE 1-DECARBOXYLASE, CHLORIDE ION, ... | | Authors: | Monteiro, D.C.F, Patel, V, Bartlett, C.P, Grant, T.D, Nozaki, S, Gowdy, J.A, Snell, E.H, Niki, H, Pearson, A.R, Webb, M.E. | | Deposit date: | 2014-03-02 | | Release date: | 2015-03-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Direct Visualisation of Strain-Induced Protein Post-Translational Modification
To be Published
|
|
5LS7
 
 | | Complex of wild type E. coli alpha aspartate decarboxylase with its processing factor PanZ | | Descriptor: | ACETYL COENZYME *A, Aspartate 1-decarboxylase, CARBON DIOXIDE, ... | | Authors: | Monteiro, D.C.F, Webb, M.E, Pearson, A.R. | | Deposit date: | 2016-08-22 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | The Mechanism of Regulation of Pantothenate Biosynthesis by the PanD-PanZAcCoA Complex Reveals an Additional Mode of Action for the Antimetabolite N-Pentyl Pantothenamide (N5-Pan).
Biochemistry, 56, 2017
|
|
4AON
 
 | |
7NEV
 
 | | Structure of the hemiacetal complex between the SARS-CoV-2 Main Protease and Leupeptin | | Descriptor: | 3C-like proteinase, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Guenther, S, Reinke, P.Y.A, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H.M, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashhour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Xavier, P.L, Ullah, N, Andaleeb, H, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Zaitsev-Doyle, J.J, Rogers, C, Gieseler, H, Melo, D, Monteiro, D.C.F, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schluenzen, F, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Sun, X, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2021-02-05 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
4AOK
 
 | |
4D7Z
 
 | | E. coli L-aspartate-alpha-decarboxylase mutant N72Q to a resolution of 1.9 Angstroms | | Descriptor: | ASPARTATE 1-DECARBOXYLASE ALPHA CHAIN, ASPARTATE 1-DECARBOXYLASE BETA CHAIN, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Bravo, J.P.K, Monteiro, D.C.F, Webb, M.E, Pearson, A.R. | | Deposit date: | 2014-12-02 | | Release date: | 2016-01-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Structure of the E. Coli L-Aspartate-Alpha-Decarboxylase Mutant N72Q to a Resolution of 1.9 Angstroms
To be Published
|
|
7ADQ
 
 | | Serial Laue crystallography structure of dehaloperoxidase B from Amphitrite ornata | | Descriptor: | Dehaloperoxidase B, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Moreno-Chicano, T.M, Ebrahim, A.E, Srajer, V, Henning, R.W, Doak, B.C, Trebbin, M, Monteiro, D.C.F, Hough, M.A. | | Deposit date: | 2020-09-15 | | Release date: | 2021-11-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Complementarity of neutron, XFEL and synchrotron crystallography for defining the structures of metalloenzymes at room temperature.
Iucrj, 9, 2022
|
|
4CRZ
 
 | | Direct visualisation of strain-induced protein prost-translational modification | | Descriptor: | ACETYL COENZYME *A, ASPARTATE 1-DECARBOXYLASE, MAGNESIUM ION, ... | | Authors: | Monteiro, D.C.F, Patel, V, Bartlett, C.P, Grant, T.D, Nozaki, S, Gowdy, J.A, Snell, E.H, Niki, H, Pearson, A.R, Webb, M.E. | | Deposit date: | 2014-03-02 | | Release date: | 2015-03-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Structure of the Pand/Panz Protein Complex Reveals Negative Feedback Regulation of Pantothenate Biosynthesis by Coenzyme A.
Chem.Biol., 22, 2015
|
|
4CS0
 
 | | Direct visualisation of strain-induced protein post-translational modification | | Descriptor: | ACETYL COENZYME *A, ASPARTATE 1-DECARBOXYLASE, MAGNESIUM ION, ... | | Authors: | Monteiro, D.C.F, Patel, V, Bartlett, C.P, Grant, T.D, Nozaki, S, Gowdy, J.A, Snell, E.H, Niki, H, Pearson, A.R, Webb, M.E. | | Deposit date: | 2014-03-02 | | Release date: | 2015-03-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Structure of the Pand/Panz Protein Complex Reveals Negative Feedback Regulation of Pantothenate Biosynthesis by Coenzyme A.
Chem.Biol., 22, 2015
|
|
6WEB
 
 | | Multi-Hit SFX using MHz XFEL sources | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Holmes, S, Darmanin, C, Abbey, B. | | Deposit date: | 2020-04-01 | | Release date: | 2021-10-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Megahertz pulse trains enable multi-hit serial femtosecond crystallography experiments at X-ray free electron lasers.
Nat Commun, 13, 2022
|
|
6WEC
 
 | | Multi-Hit SFX using MHz XFEL sources | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Holmes, S, Darmanin, C, Abbey, B. | | Deposit date: | 2020-04-01 | | Release date: | 2021-10-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Megahertz pulse trains enable multi-hit serial femtosecond crystallography experiments at X-ray free electron lasers.
Nat Commun, 13, 2022
|
|
7TUM
 
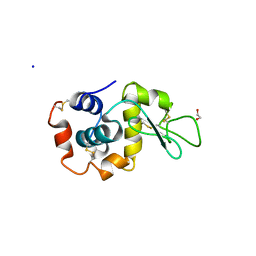 | | Multi-Hit SFX using MHz XFEL sources- first hit | | Descriptor: | 1,2-ETHANEDIOL, Lysozyme C, SODIUM ION | | Authors: | Darmanin, C, Holmes, S, Abbey, B. | | Deposit date: | 2022-02-03 | | Release date: | 2022-07-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.202 Å) | | Cite: | Megahertz pulse trains enable multi-hit serial femtosecond crystallography experiments at X-ray free electron lasers.
Nat Commun, 13, 2022
|
|
7JOR
 
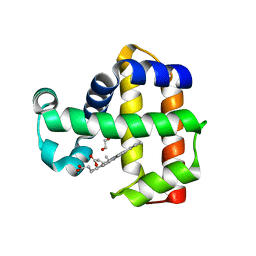 | | Neutron structure of ferric Dehaloperoxidase B | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, Dehaloperoxidase B, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Carey, L.M, Ghiladi, R.A, Meilleur, F, Myles, D.A.A. | | Deposit date: | 2020-08-07 | | Release date: | 2021-09-08 | | Last modified: | 2024-04-03 | | Method: | NEUTRON DIFFRACTION (2.05 Å) | | Cite: | Complementarity of neutron, XFEL and synchrotron crystallography for defining the structures of metalloenzymes at room temperature.
Iucrj, 9, 2022
|
|
7KCU
 
 | | Joint neutron/X-ray structure of Oxyferrous Dehaloperoxidase B | | Descriptor: | Dehaloperoxidase B, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Carey, L.M, Ghiladi, R.A, Meilleur, F, Myles, D. | | Deposit date: | 2020-10-07 | | Release date: | 2021-10-13 | | Last modified: | 2023-10-25 | | Method: | NEUTRON DIFFRACTION (2.2 Å), X-RAY DIFFRACTION | | Cite: | Complementarity of neutron, XFEL and synchrotron crystallography for defining the structures of metalloenzymes at room temperature.
Iucrj, 9, 2022
|
|
7KFM
 
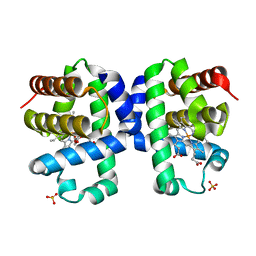 | | Room temperature oxyferrous Dehaloperoxidase B | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, Dehaloperoxidase B, OXYGEN MOLECULE, ... | | Authors: | Carey, L.M, Ghiladi, R.A. | | Deposit date: | 2020-10-14 | | Release date: | 2021-10-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Complementarity of neutron, XFEL and synchrotron crystallography for defining the structures of metalloenzymes at room temperature.
Iucrj, 9, 2022
|
|
6YNQ
 
 | | Structure of SARS-CoV-2 Main Protease bound to 2-Methyl-1-tetralone. | | Descriptor: | (2~{S})-2-methyl-3,4-dihydro-2~{H}-naphthalen-1-one, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-04-14 | | Release date: | 2020-04-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
6YVF
 
 | | Structure of SARS-CoV-2 Main Protease bound to AZD6482. | | Descriptor: | 2-[[(1R)-1-(7-methyl-2-morpholin-4-yl-4-oxidanylidene-pyrido[1,2-a]pyrimidin-9-yl)ethyl]amino]benzoic acid, 3C-like proteinase, CALCIUM ION, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-04-28 | | Release date: | 2020-05-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
6FTR
 
 | | Serial Femtosecond Crystallography at Megahertz pulse rates | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Wiedorn, M.O, Oberthuer, D, Barty, A, Chapman, H.N. | | Deposit date: | 2018-02-23 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.76000106 Å) | | Cite: | Megahertz serial crystallography.
Nat Commun, 9, 2018
|
|
7ABU
 
 | | Structure of SARS-CoV-2 Main Protease bound to RS102895 | | Descriptor: | 1'-[2-[4-(trifluoromethyl)phenyl]ethyl]spiro[1~{H}-3,1-benzoxazine-4,4'-piperidine]-2-one, 3C-like proteinase, DIMETHYL SULFOXIDE, ... | | Authors: | Guenther, S, Reinke, P.Y.A, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Lane, T.J, Dunkel, I, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-09-08 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
