1US8
 
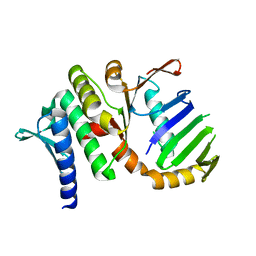 | | The Rad50 signature motif: essential to ATP binding and biological function | | Descriptor: | DNA DOUBLE-STRAND BREAK REPAIR RAD50 ATPASE | | Authors: | Moncalian, G, Lengsfeld, B, Bhaskara, V, Hopfner, K.P, Karcher, A, Alden, E, Tainer, J.A, Paull, T.T. | | Deposit date: | 2003-11-20 | | Release date: | 2003-11-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Rad50 Signature Motif: Essential to ATP Binding and Biological Function
J.Mol.Biol., 335, 2004
|
|
6H8O
 
 | |
2J6O
 
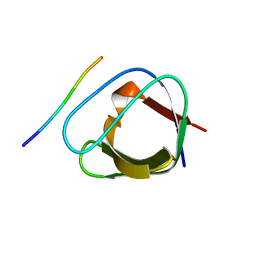 | | ATYPICAL POLYPROLINE RECOGNITION BY THE CMS N-TERMINAL SH3 DOMAIN. CMS:CD2 HETEROTRIMER | | Descriptor: | CD2-ASSOCIATED PROTEIN, T-CELL SURFACE ANTIGEN CD2 | | Authors: | Moncalian, G, Cardenes, N, Deribe, Y.L, Spinola-Amilibia, M, Dikic, I, Bravo, J. | | Deposit date: | 2006-10-02 | | Release date: | 2006-10-11 | | Last modified: | 2017-07-05 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Atypical Polyproline Recognition by the Cms N-Terminal SH3 Domain.
J.Biol.Chem., 281, 2006
|
|
2J6F
 
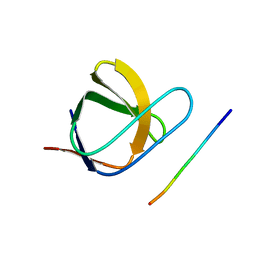 | | N-TERMINAL SH3 DOMAIN OF CMS (CD2AP HUMAN HOMOLOG) BOUND TO CBL-B PEPTIDE | | Descriptor: | CD2-ASSOCIATED PROTEIN, E3 UBIQUITIN-PROTEIN LIGASE CBL-B | | Authors: | Moncalian, G, Cardenes, N, Deribe, Y.L, Spinola-Amilibia, M, Dikic, I, Bravo, J. | | Deposit date: | 2006-09-28 | | Release date: | 2006-10-11 | | Last modified: | 2018-10-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Atypical Polyproline Recognition by the Cms N- Terminal SH3 Domain.
J.Biol.Chem., 281, 2006
|
|
2J6K
 
 | | N-TERMINAL SH3 DOMAIN OF CMS (CD2AP HUMAN HOMOLOG) | | Descriptor: | CD2-ASSOCIATED PROTEIN, SODIUM ION | | Authors: | Moncalian, G, Cardenes, N, Deribe, Y.L, Spinola-Amilibia, M, Dikic, I, Bravo, J. | | Deposit date: | 2006-09-29 | | Release date: | 2006-10-11 | | Last modified: | 2017-07-05 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Atypical Polyproline Recognition by the Cms N- Terminal SH3 Domain.
J.Biol.Chem., 281, 2006
|
|
2J7I
 
 | | ATYPICAL POLYPROLINE RECOGNITION BY THE CMS N-TERMINAL SH3 DOMAIN. CMS:CD2 HETERODIMER | | Descriptor: | CD2-ASSOCIATED PROTEIN, T-CELL SURFACE ANTIGEN CD2 | | Authors: | Moncalian, G, Cardenes, N, Deribe, Y.L, Spinola-Amilibia, M, Dikic, I, Bravo, J. | | Deposit date: | 2006-10-09 | | Release date: | 2006-11-06 | | Last modified: | 2017-07-05 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Atypical Polyproline Recognition by the Cms N-Terminal Src Homology 3 Domain.
J.Biol.Chem., 281, 2006
|
|
4PCB
 
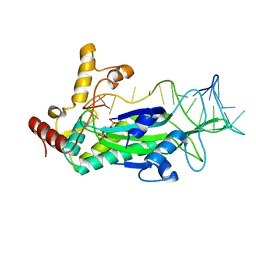 | | Conjugative Relaxase TrwC in complex with mutant OriT Dna | | Descriptor: | DNA 5'-D(P*GP*CP*AP*CP*CP*GP*AP*AP*GP*GP*TP*GP*CP*GP*TP*AP*TP*TP*CP*TP*TP*GP - 3'), PHOSPHATE ION, TrwC | | Authors: | Moncalian, G, Carballeira, J.D, de la Cruz, F, Gonzalez-Perez, B. | | Deposit date: | 2014-04-14 | | Release date: | 2014-09-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A high security double lock and key mechanism in HUH relaxases controls oriT-processing for plasmid conjugation.
Nucleic Acids Res., 42, 2014
|
|
6I89
 
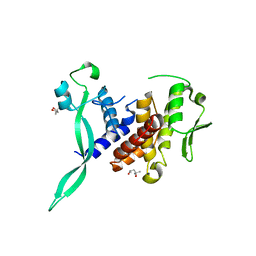 | |
7PC1
 
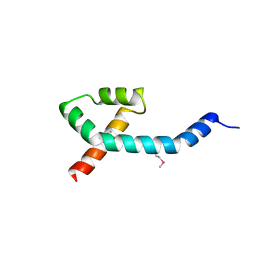 | |
3DSC
 
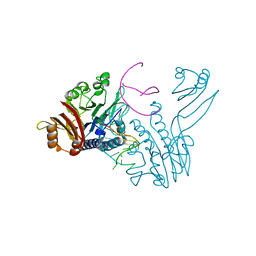 | | Crystal structure of P. furiosus Mre11 DNA synaptic complex | | Descriptor: | DNA (5'-D(P*DCP*DAP*DCP*DAP*DAP*DGP*DCP*DTP*DTP*DTP*DTP*DGP*DCP*DTP*DTP*DGP*DTP*DGP*DAP*DC)-3'), DNA double-strand break repair protein mre11 | | Authors: | Williams, R.S, Moncalian, G, Shin, D.S, Tainer, J.A. | | Deposit date: | 2008-07-11 | | Release date: | 2008-10-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mre11 dimers coordinate DNA end bridging and nuclease processing in double-strand-break repair.
Cell(Cambridge,Mass.), 135, 2008
|
|
7BCB
 
 | |
7BCA
 
 | |
6RIW
 
 | | PfaC Keto synthase-Chain length factor | | Descriptor: | MAGNESIUM ION, ORF10 | | Authors: | Santin, O, Moncalian, G. | | Deposit date: | 2019-04-25 | | Release date: | 2020-08-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure and Mechanism of the Ketosynthase-Chain Length Factor Didomain from a Prototypical Polyunsaturated Fatty Acid Synthase.
Biochemistry, 59, 2020
|
|
6SNA
 
 | |
1GL6
 
 | | Plasmid coupling protein TrwB in complex with the non-hydrolysable GTP analogue GDPNP | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ATPASE, CHLORIDE ION, ... | | Authors: | Gomis-Ruth, F.X, Moncalian, G, De La cruz, F, Coll, M. | | Deposit date: | 2001-08-28 | | Release date: | 2002-05-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Bacterial Conjugation Protein Trwb Resembles Ring Helicases and F1-ATPase
Nature, 409, 2001
|
|
1GL7
 
 | | Plasmid coupling protein TrwB in complex with the non-hydrolisable ATP-analogue ADPNP. | | Descriptor: | CHLORIDE ION, CONJUGAL TRANSFER PROTEIN TRWB, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Gomis-Ruth, F.X, Moncalian, G, De La cruz, F, Coll, M. | | Deposit date: | 2001-08-28 | | Release date: | 2002-05-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Bacterial Conjugation Protein Trwb Resembles Ring Helicases and F1-ATPase
Nature, 409, 2001
|
|
1GKI
 
 | | Plasmid coupling protein TrwB in complex with ADP and Mg2+. | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ADENOSINE-5'-DIPHOSPHATE, CONJUGAL TRANSFER PROTEIN TRWB, ... | | Authors: | Gomis-Ruth, F.X, Moncalian, G, De La cruz, F, Coll, M. | | Deposit date: | 2001-08-14 | | Release date: | 2002-05-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Bacterial Conjugation Protein Trwb Resembles Ring Helicases and F1-ATPase
Nature, 409, 2001
|
|
7BBQ
 
 | |
2AK5
 
 | | beta PIX-SH3 complexed with a Cbl-b peptide | | Descriptor: | 8-residue peptide from a signal transduction protein CBL-B, Rho guanine nucleotide exchange factor 7 | | Authors: | Jozic, D, Cardenes, N, Deribe, Y.L, Moncalian, G, Hoeller, D, Groemping, Y, Dikic, I, Rittinger, K, Bravo, J. | | Deposit date: | 2005-08-03 | | Release date: | 2005-10-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Cbl promotes clustering of endocytic adaptor proteins.
Nat.Struct.Mol.Biol., 12, 2005
|
|
2BZ8
 
 | | N-terminal Sh3 domain of CIN85 bound to Cbl-b peptide | | Descriptor: | SH3-DOMAIN KINASE BINDING PROTEIN 1, SIGNAL TRANSDUCTION PROTEIN CBL-B SH3-BINDING PROTEIN CBL-B, RING FINGER PROTEIN 56, ... | | Authors: | Cardenes, N, Moncalian, G, Bravo, J. | | Deposit date: | 2005-08-12 | | Release date: | 2005-10-05 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cbl Promotes Clustering of Endocytic Adaptor Proteins
Nat.Struct.Mol.Biol., 12, 2005
|
|
3QKU
 
 | | Mre11 Rad50 binding domain in complex with Rad50 and AMP-PNP | | Descriptor: | DNA double-strand break repair protein mre11, DNA double-strand break repair rad50 ATPase, MAGNESIUM ION, ... | | Authors: | Williams, G.J, Williams, R.S, Arvai, A, Moncalian, G, Tainer, J.A. | | Deposit date: | 2011-02-01 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | ABC ATPase signature helices in Rad50 link nucleotide state to Mre11 interface for DNA repair.
Nat.Struct.Mol.Biol., 18, 2011
|
|
3QKS
 
 | | Mre11 Rad50 binding domain bound to Rad50 | | Descriptor: | DNA double-strand break repair protein mre11, DNA double-strand break repair rad50 ATPase | | Authors: | Williams, G.J, Williams, R.S, Arvai, A, Moncalian, G, Tainer, J.A. | | Deposit date: | 2011-02-01 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | ABC ATPase signature helices in Rad50 link nucleotide state to Mre11 interface for DNA repair.
Nat.Struct.Mol.Biol., 18, 2011
|
|
3QKT
 
 | | Rad50 ABC-ATPase with adjacent coiled-coil region in complex with AMP-PNP | | Descriptor: | DNA double-strand break repair rad50 ATPase, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Williams, G.J, Williams, R.S, Arvai, A, Moncalian, G, Tainer, J.A. | | Deposit date: | 2011-02-01 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | ABC ATPase signature helices in Rad50 link nucleotide state to Mre11 interface for DNA repair.
Nat.Struct.Mol.Biol., 18, 2011
|
|
3QKR
 
 | | Mre11 Rad50 binding domain bound to Rad50 | | Descriptor: | DNA double-strand break repair protein mre11, DNA double-strand break repair rad50 ATPase, PHOSPHATE ION | | Authors: | Williams, G.J, Williams, R.S, Arvai, A, Moncalian, G, Tainer, J.A. | | Deposit date: | 2011-02-01 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | ABC ATPase signature helices in Rad50 link nucleotide state to Mre11 interface for DNA repair.
Nat.Struct.Mol.Biol., 18, 2011
|
|
1OMH
 
 | | Conjugative Relaxase TrwC in complex with OriT Dna. Metal-free structure. | | Descriptor: | DNA OLIGONUCLEOTIDE, SULFATE ION, trwC protein | | Authors: | Guasch, A, Lucas, M, Moncalian, G, Cabezas, M, Perez-Luque, R, Gomis-Ruth, F.X, de la Cruz, F, Coll, M. | | Deposit date: | 2003-02-25 | | Release date: | 2003-11-25 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Recognition and processing of the origin of transfer DNA by conjugative relaxase TrwC.
Nat.Struct.Biol., 10, 2003
|
|
