8P1R
 
 | | Bifidobacterium asteroides alpha-L-fucosidase (TT1819) catalytic mutant. | | Descriptor: | 1,2-ETHANEDIOL, Bifidobacterium asteroides alpha-L-fucosidase (TT1819) catalytic mutant, MAGNESIUM ION | | Authors: | Owen, C.D, Penner, M, Gascuena, A.N, Wu, H, Hernando, P.J, Monaco, S, Le Gall, G, Gardner, R, Ndeh, D, Urbanowicz, P.A, Spencer, D.I.R, Walsh, M.A, Angulo, J, Juge, N. | | Deposit date: | 2023-05-12 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.659 Å) | | Cite: | Exploring sequence, structure and function of microbial fucosidases from glycoside hydrolase GH29 family
To Be Published
|
|
7PMO
 
 | | Ruminococcus gnavus ATC29149 endo-beta-1,4-galactosidase (RgGH98) | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Owen, C.D, Wu, H, Crost, E.H, van Bakel, W, Gascuena, A.M, Latousakis, D, Hicks, T, Walpole, S, Urbanowicz, P.A, Ndeh, D, Monaco, S, Salom, L.S, Griffiths, R, Colvile, A, Spencer, D.I.R, Walsh, M.A, Angulo, J, Juge, N. | | Deposit date: | 2021-09-02 | | Release date: | 2022-02-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The human gut symbiont Ruminococcus gnavus shows specificity to blood group A antigen during mucin glycan foraging: Implication for niche colonisation in the gastrointestinal tract.
Plos Biol., 19, 2021
|
|
7Q20
 
 | | Ruminococcus gnavus ATC29149 endo-beta-1,4-galactosidase (RgGH98) in complex with blood group A trisaccharide | | Descriptor: | CALCIUM ION, MAGNESIUM ION, Ruminococcus gnavus endogalactosidase GH98, ... | | Authors: | Owen, C.D, Wu, H, Crost, E.H, van Bakel, W, Gascuena, A.M, Latousakis, D, Hicks, T, Walpole, S, Urbanowicz, P.A, Ndeh, D, Monaco, S, Salom, L.S, Griffiths, R, Colvile, A, Spencer, D.I.R, Walsh, M.A, Angulo, J, Juge, N. | | Deposit date: | 2021-10-22 | | Release date: | 2022-02-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The human gut symbiont Ruminococcus gnavus shows specificity to blood group A antigen during mucin glycan foraging: Implication for niche colonisation in the gastrointestinal tract.
Plos Biol., 19, 2021
|
|
7Q1W
 
 | | Ruminococcus gnavus ATC29149 endo-beta-1,4-galactosidase (RgGH98) E411A in complex with blood group A (BgA II) tetrasaccharide | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Owen, C.D, Wu, H, Crost, E.H, van Bakel, W, Gascuena, A.M, Latousakis, D, Hicks, T, Walpole, S, Urbanowicz, P.A, Ndeh, D, Monaco, S, Salom, L.S, Griffiths, R, Colvile, A, Spencer, D.I.R, Walsh, M.A, Angulo, J, Juge, N. | | Deposit date: | 2021-10-21 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The human gut symbiont Ruminococcus gnavus shows specificity to blood group A antigen during mucin glycan foraging: Implication for niche colonisation in the gastrointestinal tract.
Plos Biol., 19, 2021
|
|
1E6J
 
 | | Crystal structure of HIV-1 capsid protein (p24) in complex with Fab13B5 | | Descriptor: | CAPSID PROTEIN P24, IMMUNOGLOBULIN | | Authors: | Berthet-Colominas, C, Monaco, S, Novelli, A, Sibai, G, Mallet, F, Cusack, S. | | Deposit date: | 2000-08-18 | | Release date: | 2000-11-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Mutual Conformational Adaptations in Antigen and Antibody Upon Complex Formation between an Fab and HIV-1 Capsid Protein P24
Structure, 8, 2000
|
|
5H63
 
 | | Structure of Transferase mutant-C23S,C199S | | Descriptor: | MANGANESE (II) ION, Transferase, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE | | Authors: | Park, J.B, Yoo, Y, Kim, J. | | Deposit date: | 2016-11-10 | | Release date: | 2017-12-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural basis for arginine glycosylation of host substrates by bacterial effector proteins.
Nat Commun, 9, 2018
|
|
5H62
 
 | | Structure of Transferase mutant-C23S,C199S | | Descriptor: | 1,2-ETHANEDIOL, MANGANESE (II) ION, Transferase, ... | | Authors: | Park, J.B, Yoo, Y, Kim, J. | | Deposit date: | 2016-11-10 | | Release date: | 2017-12-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structural basis for arginine glycosylation of host substrates by bacterial effector proteins.
Nat Commun, 9, 2018
|
|
5H60
 
 | | Structure of Transferase mutant-C23S,C199S | | Descriptor: | MANGANESE (II) ION, Transferase, URIDINE-5'-DIPHOSPHATE | | Authors: | Park, J.B, Yoo, Y, Kim, J. | | Deposit date: | 2016-11-10 | | Release date: | 2017-12-20 | | Last modified: | 2018-10-31 | | Method: | X-RAY DIFFRACTION (3.64 Å) | | Cite: | Structural basis for arginine glycosylation of host substrates by bacterial effector proteins.
Nat Commun, 9, 2018
|
|
5H61
 
 | | Structure of Transferase mutant-C23S,C199S | | Descriptor: | Transferase | | Authors: | Park, J.B, Yoo, Y, Kim, J. | | Deposit date: | 2016-11-10 | | Release date: | 2017-12-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural basis for arginine glycosylation of host substrates by bacterial effector proteins.
Nat Commun, 9, 2018
|
|
5H5Y
 
 | | Structure of Transferase mutant-C23S,C199S | | Descriptor: | Non-LEE encoded effector protein NleB | | Authors: | Park, J.B, Yoo, Y, Kim, J. | | Deposit date: | 2016-11-10 | | Release date: | 2017-12-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for arginine glycosylation of host substrates by bacterial effector proteins.
Nat Commun, 9, 2018
|
|
6Z3C
 
 | | High resolution structure of RgNanOx | | Descriptor: | CITRATE ANION, Gfo/Idh/MocA family oxidoreductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Naismith, J.H, Lee, M.O. | | Deposit date: | 2020-05-19 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Uncovering a novel molecular mechanism for scavenging sialic acids in bacteria.
J.Biol.Chem., 295, 2020
|
|
6Z3B
 
 | | Low resolution structure of RgNanOx | | Descriptor: | CITRIC ACID, Gfo/Idh/MocA family oxidoreductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Naismith, J.H, Lee, M. | | Deposit date: | 2020-05-19 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Uncovering a novel molecular mechanism for scavenging sialic acids in bacteria.
J.Biol.Chem., 295, 2020
|
|
8HLF
 
 | | Crystal structure of DddK-DMSOP complex | | Descriptor: | 3-[dimethyl(oxidanyl)-$l^{4}-sulfanyl]propanoic acid, MANGANESE (II) ION, Novel protein with potential Cupin domain | | Authors: | Peng, M, Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2022-11-30 | | Release date: | 2023-10-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | DMSOP-cleaving enzymes are diverse and widely distributed in marine microorganisms.
Nat Microbiol, 8, 2023
|
|
8HLE
 
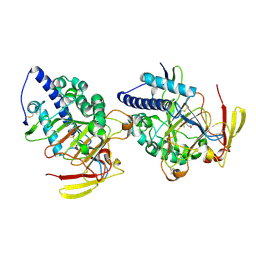 | | Structure of DddY-DMSOP complex | | Descriptor: | 3-[dimethyl(oxidanyl)-$l^{4}-sulfanyl]propanoic acid, DMSP lyase DddY, ZINC ION | | Authors: | Peng, M, Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2022-11-30 | | Release date: | 2023-10-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | DMSOP-cleaving enzymes are diverse and widely distributed in marine microorganisms.
Nat Microbiol, 8, 2023
|
|
8BHD
 
 | | N-terminal domain of Plasmodium berghei glutamyl-tRNA synthetase (Tbxo4 derivative crystal structure) | | Descriptor: | GLYCEROL, Glutamate--tRNA ligase, SULFATE ION, ... | | Authors: | Benas, P, Jaramillo Ponce, J.R, Legrand, P, Frugier, M, Sauter, C. | | Deposit date: | 2022-10-31 | | Release date: | 2023-01-25 | | Last modified: | 2023-02-08 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | Solution X-ray scattering highlights discrepancies in Plasmodium multi-aminoacyl-tRNA synthetase complexes.
Protein Sci., 32, 2023
|
|
6ER3
 
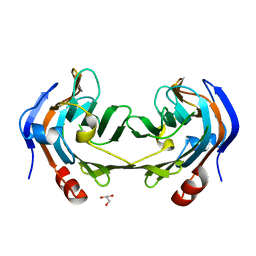 | | Ruminococcus gnavus IT-sialidase CBM40 bound to alpha2,3 sialyllactose | | Descriptor: | BNR/Asp-box repeat protein, GLYCEROL, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose | | Authors: | Owen, C.D, Tailford, L.E, Taylor, G.L, Juge, N. | | Deposit date: | 2017-10-16 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Unravelling the specificity and mechanism of sialic acid recognition by the gut symbiont Ruminococcus gnavus.
Nat Commun, 8, 2017
|
|
6ER4
 
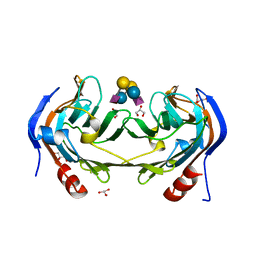 | | Ruminococcus gnavus IT-sialidase CBM40 bound to alpha2,6 sialyllactose | | Descriptor: | BNR/Asp-box repeat protein, GLYCEROL, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Owen, C.D, Tailford, L.E, Taylor, G.L, Juge, N. | | Deposit date: | 2017-10-16 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Unravelling the specificity and mechanism of sialic acid recognition by the gut symbiont Ruminococcus gnavus.
Nat Commun, 8, 2017
|
|
6ER2
 
 | |
6FJ2
 
 | |
6FJ9
 
 | |
6FJ4
 
 | |
6FJ6
 
 | |
6FID
 
 | | Bovine trypsin solved by S-SAD on ID30B | | Descriptor: | BENZAMIDINE, CALCIUM ION, Cationic trypsin, ... | | Authors: | McCarthy, A.A, Mueller-Dieckmann, C. | | Deposit date: | 2018-01-18 | | Release date: | 2018-02-07 | | Last modified: | 2018-08-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | ID30B - a versatile beamline for macromolecular crystallography experiments at the ESRF.
J Synchrotron Radiat, 25, 2018
|
|
6FJ8
 
 | |
6TR3
 
 | | Ruminococcus gnavus GH29 fucosidase E1_10125 in complex with fucose | | Descriptor: | CALCIUM ION, F5/8 type C domain-containing protein, MAGNESIUM ION, ... | | Authors: | Owen, C.D, Wu, H, Crost, E, Colvile, A, Juge, N, Walsh, M.A. | | Deposit date: | 2019-12-17 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Fucosidases from the human gut symbiont Ruminococcus gnavus.
Cell.Mol.Life Sci., 78, 2021
|
|
