8KME
 
 | |
4KIV
 
 | |
2C00
 
 | |
3KIV
 
 | | RECOMBINANT KRINGLE IV-10/M66 VARIANT OF HUMAN APOLIPOPROTEIN(A) | | Descriptor: | 6-AMINOHEXANOIC ACID, APOLIPOPROTEIN | | Authors: | Mochalkin, I, Tulinsky, A, Scanu, A. | | Deposit date: | 1998-09-08 | | Release date: | 1999-05-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Recombinant kringle IV-10 modules of human apolipoprotein(a): structure, ligand binding modes, and biological relevance.
Biochemistry, 38, 1999
|
|
7KME
 
 | |
1KIV
 
 | |
2J9G
 
 | |
2V0L
 
 | | Characterization of Substrate Binding and Catalysis of the Potential Antibacterial Target N-acetylglucosamine-1-phosphate Uridyltransferase (GlmU) | | Descriptor: | BIFUNCTIONAL PROTEIN GLMU, SULFATE ION, TETRAETHYLENE GLYCOL, ... | | Authors: | Mochalkin, I, Lightle, S, Ohren, J.F, Chirgadze, N.Y. | | Deposit date: | 2007-05-14 | | Release date: | 2008-01-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Characterization of Substrate Binding and Catalysis in the Potential Antibacterial Target N-Acetylglucosamine-1-Phosphate Uridyltransferase (Glmu).
Protein Sci., 16, 2007
|
|
2V0H
 
 | | Characterization of Substrate Binding and Catalysis of the Potential Antibacterial Target N-acetylglucosamine-1-phosphate Uridyltransferase (GlmU) | | Descriptor: | BIFUNCTIONAL PROTEIN GLMU, COBALT (II) ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Mochalkin, I, Lightle, S, Ohren, J.F, Chirgadze, N.Y. | | Deposit date: | 2007-05-14 | | Release date: | 2008-01-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Characterization of Substrate Binding and Catalysis in the Potential Antibacterial Target N-Acetylglucosamine-1-Phosphate Uridyltransferase (Glmu).
Protein Sci., 16, 2007
|
|
2V0I
 
 | | Characterization of Substrate Binding and Catalysis of the Potential Antibacterial Target N-acetylglucosamine-1-phosphate Uridyltransferase (GlmU) | | Descriptor: | BIFUNCTIONAL PROTEIN GLMU, DI(HYDROXYETHYL)ETHER, SULFATE ION, ... | | Authors: | Mochalkin, I, Lightle, S, Ohren, J.F, Chirgadze, N.Y. | | Deposit date: | 2007-05-14 | | Release date: | 2008-01-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Characterization of Substrate Binding and Catalysis in the Potential Antibacterial Target N-Acetylglucosamine-1-Phosphate Uridyltransferase (Glmu).
Protein Sci., 16, 2007
|
|
2V0J
 
 | | Characterization of Substrate Binding and Catalysis of the Potential Antibacterial Target N-acetylglucosamine-1-phosphate Uridyltransferase (GlmU) | | Descriptor: | 5,6-DIHYDROURIDINE-5'-MONOPHOSPHATE, BIFUNCTIONAL PROTEIN GLMU, MAGNESIUM ION, ... | | Authors: | Mochalkin, I, Lightle, S, Ohren, J.F, Chirgadze, N.Y. | | Deposit date: | 2007-05-14 | | Release date: | 2008-01-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Characterization of Substrate Binding and Catalysis in the Potential Antibacterial Target N-Acetylglucosamine-1-Phosphate Uridyltransferase (Glmu).
Protein Sci., 16, 2007
|
|
2V0K
 
 | | Characterization of Substrate Binding and Catalysis of the Potential Antibacterial Target N-acetylglucosamine-1-phosphate Uridyltransferase (GlmU) | | Descriptor: | BIFUNCTIONAL PROTEIN GLMU, SULFATE ION, TETRAETHYLENE GLYCOL, ... | | Authors: | Mochalkin, I, Lightle, S, Ohren, J.F, Chirgadze, N.Y. | | Deposit date: | 2007-05-14 | | Release date: | 2008-01-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Characterization of Substrate Binding and Catalysis in the Potential Antibacterial Target N-Acetylglucosamine-1-Phosphate Uridyltransferase (Glmu).
Protein Sci., 16, 2007
|
|
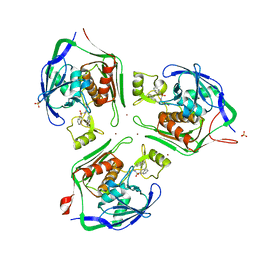
2VES
 
 | |
2VD4
 
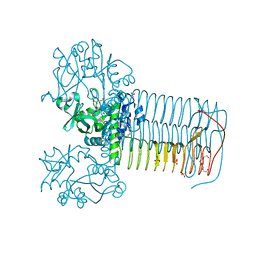 | | Structure of small-molecule inhibitor of Glmu from Haemophilus influenzae reveals an allosteric binding site | | Descriptor: | 4-chloro-N-(3-methoxypropyl)-N-[(3S)-1-(2-phenylethyl)piperidin-3-yl]benzamide, BIFUNCTIONAL PROTEIN GLMU, MAGNESIUM ION, ... | | Authors: | Mochalkin, I, Lightle, S, McDowell, L. | | Deposit date: | 2007-09-28 | | Release date: | 2008-01-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a Small-Molecule Inhibitor Complexed with Glmu from Haemophilus Influenzae Reveals an Allosteric Binding Site.
Protein Sci., 17, 2008
|
|
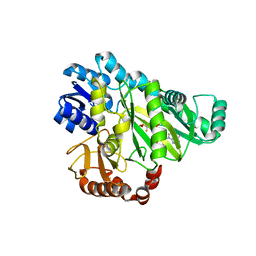
2VR1
 
 | |
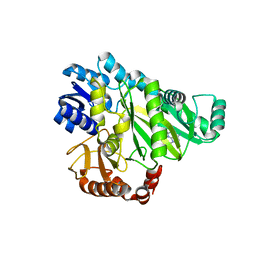
2W6Z
 
 | |
2W6M
 
 | |
2W6P
 
 | |
2W70
 
 | |
2W6N
 
 | |
2VPQ
 
 | |
2VQD
 
 | |
2W6O
 
 | | Crystal structure of Biotin carboxylase from E. coli in complex with 4-Amino-7,7-dimethyl-7,8-dihydro-quinazolinone fragment | | Descriptor: | 4-amino-7,7-dimethyl-7,8-dihydroquinazolin-5(6H)-one, BIOTIN CARBOXYLASE, CHLORIDE ION | | Authors: | Mochalkin, I, Miller, J.R. | | Deposit date: | 2008-12-18 | | Release date: | 2009-05-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of Antibacterial Biotin Carboxylase Inhibitors by Virtual Screening and Fragment-Based Approaches.
Acs Chem.Biol., 4, 2009
|
|
2W6Q
 
 | | Crystal structure of Biotin carboxylase from E. coli in complex with the triazine-2,4-diamine fragment | | Descriptor: | 6-(2-phenoxyethoxy)-1,3,5-triazine-2,4-diamine, BIOTIN CARBOXYLASE, CHLORIDE ION | | Authors: | Mochalkin, I, Miller, J.R. | | Deposit date: | 2008-12-18 | | Release date: | 2009-05-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery of Antibacterial Biotin Carboxylase Inhibitors by Virtual Screening and Fragment-Based Approaches.
Acs Chem.Biol., 4, 2009
|
|
2W71
 
 | |
