1G7X
 
 | | ASPARTATE AMINOTRANSFERASE ACTIVE SITE MUTANT N194A/R292L/R386L | | Descriptor: | ASPARTATE AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Mizuguchi, H, Hayashi, H, Okada, K, Miyahara, I, Hirotsu, K, Kagamiyama, H. | | Deposit date: | 2000-11-15 | | Release date: | 2000-11-29 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Strain is more important than electrostatic interaction in controlling the pKa of the catalytic group in aspartate aminotransferase.
Biochemistry, 40, 2001
|
|
1G4V
 
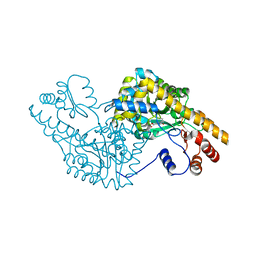
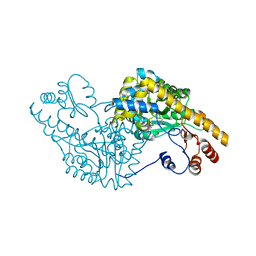 | | ASPARTATE AMINOTRANSFERASE ACTIVE SITE MUTANT N194A/Y225F | | Descriptor: | ASPARTATE AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Mizuguchi, H, Hayashi, H, Okada, K, Miyahara, I, Hirotsu, K, Kagamiyama, H. | | Deposit date: | 2000-10-28 | | Release date: | 2000-11-22 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Strain is more important than electrostatic interaction in controlling the pKa of the catalytic group in aspartate aminotransferase.
Biochemistry, 40, 2001
|
|
1G7W
 
 | | ASPARTATE AMINOTRANSFERASE ACTIVE SITE MUTANT N194A/R386L | | Descriptor: | ASPARTATE AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Mizuguchi, H, Hayashi, H, Okada, K, Miyahara, I, Hirotsu, K, Kagamiyama, H. | | Deposit date: | 2000-11-15 | | Release date: | 2000-11-29 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Strain is more important than electrostatic interaction in controlling the pKa of the catalytic group in aspartate aminotransferase.
Biochemistry, 40, 2001
|
|
1G4X
 
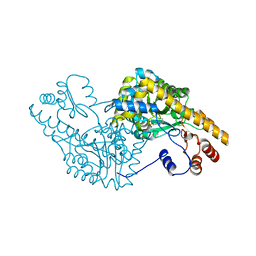 | | ASPARTATE AMINOTRANSFERASE ACTIVE SITE MUTANT N194A/R292L | | Descriptor: | ASPARTATE AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Mizuguchi, H. | | Deposit date: | 2000-10-29 | | Release date: | 2000-11-22 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Strain is more important than electrostatic interaction in controlling the pKa of the catalytic group in aspartate aminotransferase.
Biochemistry, 40, 2001
|
|
1GEW
 
 | | CRYSTAL STRUCTURE OF HISTIDINOL-PHOSPHATE AMINOTRANSFERASE COMPLEXED WITH PYRIDOXAL 5'-PHOSPHATE | | Descriptor: | HISTIDINOL-PHOSPHATE AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Haruyama, K, Nakai, T, Miyahara, I, Hirotsu, K, Mizuguchi, H, Hayashi, H, Kagamiyama, H. | | Deposit date: | 2000-11-30 | | Release date: | 2001-04-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of Escherichia coli histidinol-phosphate aminotransferase and its complexes with histidinol-phosphate and N-(5'-phosphopyridoxyl)-L-glutamate: double substrate recognition of the enzyme.
Biochemistry, 40, 2001
|
|
1GEY
 
 | | CRYSTAL STRUCTURE OF HISTIDINOL-PHOSPHATE AMINOTRANSFERASE COMPLEXED WITH N-(5'-PHOSPHOPYRIDOXYL)-L-GLUTAMATE | | Descriptor: | 4-[(1,3-DICARBOXY-PROPYLAMINO)-METHYL]-3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDINIUM, HISTIDINOL-PHOSPHATE AMINOTRANSFERASE | | Authors: | Haruyama, K, Nakai, T, Miyahara, I, Hirotsu, K, Mizuguchi, H, Hayashi, H, Kagamiyama, H. | | Deposit date: | 2000-11-30 | | Release date: | 2001-04-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of Escherichia coli histidinol-phosphate aminotransferase and its complexes with histidinol-phosphate and N-(5'-phosphopyridoxyl)-L-glutamate: double substrate recognition of the enzyme.
Biochemistry, 40, 2001
|
|
1GEX
 
 | | CRYSTAL STRUCTURE OF HISTIDINOL-PHOSPHATE AMINOTRANSFERASE COMPLEXED WITH HISTIDINOL-PHOSPHATE | | Descriptor: | HISTIDINOL-PHOSPHATE AMINOTRANSFERASE, PHOSPHORIC ACID MONO-[2-AMINO-3-(3H-IMIDAZOL-4-YL)-PROPYL]ESTER, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Haruyama, K, Nakai, T, Miyahara, I, Hirotsu, K, Mizuguchi, H, Hayashi, H, Kagamiyama, H. | | Deposit date: | 2000-11-30 | | Release date: | 2001-04-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of Escherichia coli histidinol-phosphate aminotransferase and its complexes with histidinol-phosphate and N-(5'-phosphopyridoxyl)-L-glutamate: double substrate recognition of the enzyme.
Biochemistry, 40, 2001
|
|
1IX6
 
 | | Aspartate Aminotransferase Active Site Mutant V39F | | Descriptor: | Aspartate Aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Hayashi, H, Mizuguchi, H, Miyahara, I, Nakajima, Y, Hirotsu, K, Kagamiyama, H. | | Deposit date: | 2002-06-14 | | Release date: | 2002-07-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Conformational change in aspartate aminotransferase on substrate binding induces strain in the catalytic group and enhances catalysis
J.BIOL.CHEM., 278, 2003
|
|
1IX8
 
 | | Aspartate Aminotransferase Active Site Mutant V39F/N194A | | Descriptor: | Aspartate Aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Hayashi, H, Mizuguchi, H, Miyahara, I, Nakajima, Y, Hirotsu, K, Kagamiyama, H. | | Deposit date: | 2002-06-14 | | Release date: | 2002-07-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Conformational change in aspartate aminotransferase on substrate binding induces strain in the catalytic group and enhances catalysis
J.BIOL.CHEM., 278, 2003
|
|
1IX7
 
 | | Aspartate Aminotransferase Active Site Mutant V39F maleate complex | | Descriptor: | Aspartate Aminotransferase, MALEIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Hayashi, H, Mizuguchi, H, Miyahara, I, Nakajima, Y, Hirotsu, K, Kagamiyama, H. | | Deposit date: | 2002-06-14 | | Release date: | 2002-07-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Conformational change in aspartate aminotransferase on substrate binding induces strain in the catalytic group and enhances catalysis
J.BIOL.CHEM., 278, 2003
|
|
2BIF
 
 | | 6-PHOSPHOFRUCTO-2-KINASE/FRUCTOSE-2,6-BISPHOSPHATASE H256A MUTANT WITH F6P IN PHOSPHATASE ACTIVE SITE | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Yuen, M.H, Hasemann, C.A. | | Deposit date: | 1998-10-26 | | Release date: | 1999-02-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the H256A mutant of rat testis fructose-6-phosphate,2-kinase/fructose-2,6-bisphosphatase. Fructose 6-phosphate in the active site leads to mechanisms for both mutant and wild type bisphosphatase activities.
J.Biol.Chem., 274, 1999
|
|
3X0V
 
 | | Structure of L-lysine oxidase | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, L-lysine oxidase | | Authors: | Sano, T, Uchida, Y, Amano, M, Kawaguchi, T, Kondo, H, Inagaki, K, Imada, K. | | Deposit date: | 2014-10-22 | | Release date: | 2015-04-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Recombinant expression, molecular characterization and crystal structure of antitumor enzyme, l-lysine alpha-oxidase from Trichoderma viride.
J.Biochem., 157, 2015
|
|
1UIM
 
 | |
1UIN
 
 | |
1V2F
 
 | |
1V2D
 
 | |
1V7C
 
 | |
1V2E
 
 | |
1VB3
 
 | |
1KA9
 
 | |
3BIF
 
 | | 6-PHOSPHOFRUCTO-2-KINASE/FRUCTOSE-2,6-BISPHOSPHATASE EMPTY 6-PF-2K ACTIVE SITE | | Descriptor: | PHOSPHATE ION, PROTEIN (6-PHOSPHOFRUCTO-2-KINASE/ FRUCTOSE-2,6-BISPHOSPHATASE), SUCCINIC ACID, ... | | Authors: | Yuen, M.H, Hasemann, C.A. | | Deposit date: | 1999-05-05 | | Release date: | 1999-09-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A switch in the kinase domain of rat testis 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase.
Biochemistry, 38, 1999
|
|
