1B2T
 
 | |
1F2L
 
 | |
1QX2
 
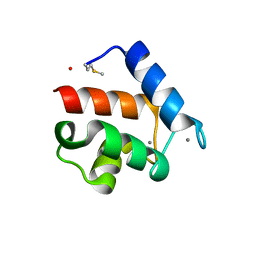 | | X-ray Structure of Calcium-loaded Calbindomodulin (A Calbindin D9k Re-engineered to Undergo a Conformational Opening) at 1.44 A Resolution | | Descriptor: | CALCIUM ION, Vitamin D-dependent calcium-binding protein, intestinal, ... | | Authors: | Bunick, C.G, Nelson, M.R, Mangahas, S, Mizoue, L.S, Bunick, G.J, Chazin, W.J. | | Deposit date: | 2003-09-04 | | Release date: | 2004-05-25 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Designing Sequence to Control Protein Function in an EF-Hand Protein
J.Am.Chem.Soc., 126, 2004
|
|
3MFA
 
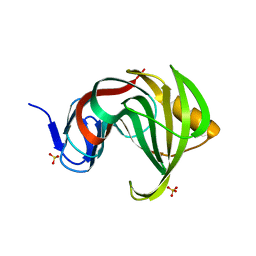 | | Computationally designed endo-1,4-beta-xylanase | | Descriptor: | Endo-1,4-beta-xylanase, SULFATE ION | | Authors: | Morin, A, Harp, J.M. | | Deposit date: | 2010-04-01 | | Release date: | 2010-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Computational design of an endo-1,4-{beta}-xylanase ligand binding site.
Protein Eng.Des.Sel., 24, 2011
|
|
3MF9
 
 | | Computationally designed endo-1,4-beta-xylanase | | Descriptor: | Endo-1,4-beta-xylanase, SULFATE ION | | Authors: | Morin, A, Harp, J.M. | | Deposit date: | 2010-04-01 | | Release date: | 2010-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Computational design of an endo-1,4-{beta}-xylanase ligand binding site.
Protein Eng.Des.Sel., 24, 2011
|
|
3MFC
 
 | | Computationally designed end0-1,4-beta,xylanase | | Descriptor: | Endo-1,4-beta-xylanase, SULFATE ION | | Authors: | Morin, A, Harp, J.M. | | Deposit date: | 2010-04-01 | | Release date: | 2010-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Computational design of an endo-1,4-{beta}-xylanase ligand binding site.
Protein Eng.Des.Sel., 24, 2011
|
|
3MF6
 
 | | Computationally designed endo-1,4-beta-xylanase | | Descriptor: | Endo-1,4-beta-xylanase, SULFATE ION | | Authors: | Morin, A, Harp, J.M. | | Deposit date: | 2010-04-01 | | Release date: | 2010-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Computational design of an endo-1,4-{beta}-xylanase ligand binding site.
Protein Eng.Des.Sel., 24, 2011
|
|
