8XXA
 
 | |
8XX9
 
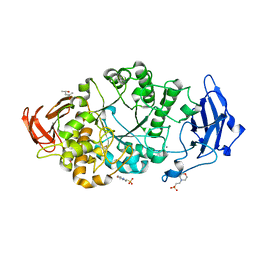 | | Rhodothermus marinus alpha-amylase RmGH13_47A CBM48-A-B-C domains | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Tonozuka, T. | | Deposit date: | 2024-01-18 | | Release date: | 2024-02-07 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural basis for the recognition of alpha-1,6-branched alpha-glucan by GH13_47 alpha-amylase from Rhodothermus marinus
Proteins, 2024
|
|
7XOI
 
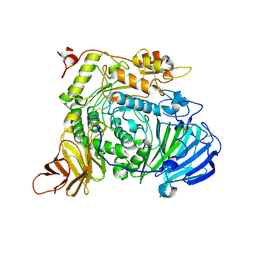 | | Aspergillus sojae alpha-glucosidase AsojAgdL in complex with trehalose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, SODIUM ION, ... | | Authors: | Tonozuka, T. | | Deposit date: | 2022-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for proteolytic processing of Aspergillus sojae alpha-glucosidase L with strong transglucosylation activity.
J.Struct.Biol., 214, 2022
|
|
8I2Q
 
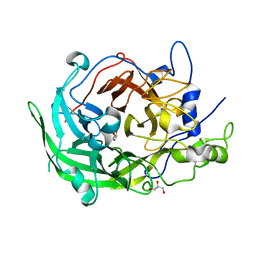 | | Beijerinckia indica beta-fructosyltransferase variant H395R/F473Y | | Descriptor: | Beta-fructosyltransferase, GLYCEROL | | Authors: | Tonozuka, T. | | Deposit date: | 2023-01-15 | | Release date: | 2023-06-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Characterization and alteration of product specificity of Beijerinckia indica subsp. indica beta-fructosyltransferase.
Biosci.Biotechnol.Biochem., 87, 2023
|
|
8I2R
 
 | |
1WMR
 
 | | Crystal Structure of Isopullulanase from Aspergillus niger ATCC 9642 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Isopullulanase | | Authors: | Mizuno, M, Tonozuka, T, Miyasaka, Y, Akeboshi, H, Kamitori, S, Nishikawa, A, Sakano, Y. | | Deposit date: | 2004-07-15 | | Release date: | 2005-07-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Aspergillus niger Isopullulanase, a Member of Glycoside Hydrolase Family 49
J.Mol.Biol., 376, 2008
|
|
1X0C
 
 | | Improved Crystal Structure of Isopullulanase from Aspergillus niger ATCC 9642 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Isopullulanase | | Authors: | Mizuno, M, Tonozuka, T, Yamamura, A, Miyasaka, Y, Akeboshi, H, Kamitori, S, Nishikawa, A, Sakano, Y. | | Deposit date: | 2005-03-17 | | Release date: | 2006-06-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Aspergillus niger Isopullulanase, a Member of Glycoside Hydrolase Family 49
J.Mol.Biol., 376, 2008
|
|
