1J18
 
 | | Crystal Structure of a Beta-Amylase from Bacillus cereus var. mycoides Cocrystallized with Maltose | | Descriptor: | ACETIC ACID, Beta-amylase, CALCIUM ION, ... | | Authors: | Miyake, H, Kurisu, G, Kusunoki, M, Nishimura, S, Kitamura, S, Nitta, Y. | | Deposit date: | 2002-12-02 | | Release date: | 2003-05-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of a Catalytic Site Mutant of beta-Amylase from Bacillus cereus var. mycoides Cocrystallized with Maltopentaose
BIOCHEMISTRY, 42, 2003
|
|
1ITC
 
 | | Beta-Amylase from Bacillus cereus var. mycoides Complexed with Maltopentaose | | Descriptor: | ACETIC ACID, Beta-Amylase, CALCIUM ION, ... | | Authors: | Miyake, H, Kurisu, G, Kusunoki, M, Nishimura, S, Kitamura, S, Nitta, Y. | | Deposit date: | 2002-01-17 | | Release date: | 2003-05-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of a Catalytic Site Mutant of beta-Amylase from Bacillus cereus var. mycoides Cocrystallized with Maltopentaose
BIOCHEMISTRY, 42, 2003
|
|
5GHL
 
 | | Crystal structure Analysis of the starch-binding domain of glucoamylase from Aspergillus niger | | Descriptor: | GLYCEROL, Glucoamylase, SULFATE ION | | Authors: | Miyake, H, Suyama, Y, Muraki, N, Kusunoki, M, Tanaka, A. | | Deposit date: | 2016-06-20 | | Release date: | 2017-10-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the starch-binding domain of glucoamylase from Aspergillus niger.
Acta Crystallogr.,Sect.F, 73, 2017
|
|
1OMY
 
 | | Crystal Structure of a Recombinant alpha-insect Toxin BmKaIT1 from the scorpion Buthus martensii Karsch | | Descriptor: | ACETIC ACID, Alpha-neurotoxin TX12, CHLORIDE ION | | Authors: | Huang, Y, Huang, Q, Chen, H, Tang, Y, Miyake, H, Kusunoki, M. | | Deposit date: | 2003-02-26 | | Release date: | 2003-09-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallization and preliminary crystallographic study of rBmKalphaIT1, a recombinant alpha-insect toxin from the scorpion Buthus martensii Karsch.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1OX1
 
 | | crystal structure of the bovine trypsin complex with a synthetic 11 peptide inhibitor | | Descriptor: | 11-mer peptide, CALCIUM ION, Trypsinogen, ... | | Authors: | Wu, G, Huang, Y, Zhu, G, Huang, Q, Tang, Y, Miyake, H, Kusunoki, M. | | Deposit date: | 2003-03-31 | | Release date: | 2004-05-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | crystal structure of the bovine trypsin complex with a synthetic 11 peptide inhibitor
To be published
|
|
3A4A
 
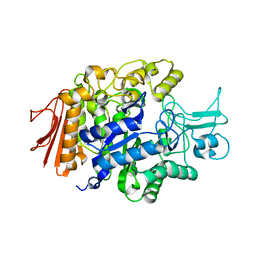 | | Crystal structure of isomaltase from Saccharomyces cerevisiae | | Descriptor: | CALCIUM ION, Oligo-1,6-glucosidase, alpha-D-glucopyranose | | Authors: | Yamamoto, K, Miyake, H, Kusunoki, M, Osaki, S. | | Deposit date: | 2009-07-01 | | Release date: | 2010-07-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of isomaltase from Saccharomyces cerevisiae and in complex with its competitive inhibitor maltose
Febs J., 277, 2010
|
|
3A47
 
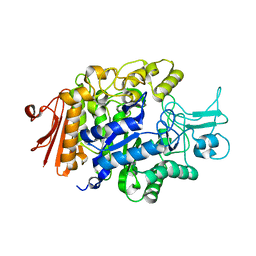 | |
1WOK
 
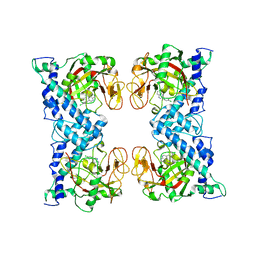 | | Crystal structure of catalytic domain of human poly(ADP-ribose) polymerase complexed with a quinoxaline-type inhibitor | | Descriptor: | 3-(4-CHLOROPHENYL)QUINOXALINE-5-CARBOXAMIDE, Poly [ADP-ribose] polymerase-1 | | Authors: | Iwashita, A, Hattori, K, Yamamoto, H, Ishida, J, Kido, Y, Kamijo, K, Murano, K, Miyake, H, Kinoshita, T, Warizaya, M, Ohkubo, M, Matsuoka, N, Mutoh, S. | | Deposit date: | 2004-08-20 | | Release date: | 2005-03-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Discovery of quinazolinone and quinoxaline derivatives as potent and selective poly(ADP-ribose) polymerase-1/2 inhibitors.
Febs Lett., 579, 2005
|
|
3AJ7
 
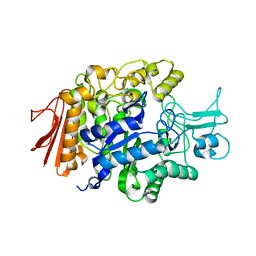 | | Crystal Structure of isomaltase from Saccharomyces cerevisiae | | Descriptor: | CALCIUM ION, Oligo-1,6-glucosidase | | Authors: | Yamamoto, K, Miyake, H, Kusunoki, M, Osaki, S. | | Deposit date: | 2010-05-26 | | Release date: | 2010-08-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structures of isomaltase from Saccharomyces cerevisiae and in complex with its competitive inhibitor maltose
Febs J., 277, 2010
|
|
3AXH
 
 | | Crystal structure of isomaltase in complex with isomaltose | | Descriptor: | CALCIUM ION, Oligo-1,6-glucosidase IMA1, alpha-D-glucopyranose-(1-6)-alpha-D-glucopyranose | | Authors: | Yamamoto, K, Miyake, H, Kusunoki, M, Osaki, S. | | Deposit date: | 2011-04-06 | | Release date: | 2011-10-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Steric hindrance by 2 amino acid residues determines the substrate specificity of isomaltase from Saccharomyces cerevisiae
J.Biosci.Bioeng., 112, 2011
|
|
3AXI
 
 | | Crystal structure of isomaltase in complex with maltose | | Descriptor: | CALCIUM ION, Oligo-1,6-glucosidase IMA1, alpha-D-glucopyranose | | Authors: | Yamamoto, K, Miyake, H, Kusunoki, M, Osaki, S. | | Deposit date: | 2011-04-06 | | Release date: | 2011-10-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Steric hindrance by 2 amino acid residues determines the substrate specificity of isomaltase from Saccharomyces cerevisiae
J.Biosci.Bioeng., 112, 2011
|
|
1J10
 
 | | beta-amylase from Bacillus cereus var. mycoides in complex with GGX | | Descriptor: | Beta-amylase, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-xylopyranose, ... | | Authors: | Oyama, T, Miyake, H, Kusunoki, M, Nitta, Y. | | Deposit date: | 2002-11-25 | | Release date: | 2003-06-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of beta-Amylase from Bacillus cereus var. mycoides in Complexes with Substrate Analogs and Affinity-Labeling Reagents
J.BIOCHEM.(TOKYO), 133, 2003
|
|
1IEI
 
 | | CRYSTAL STRUCTURE OF HUMAN ALDOSE REDUCTASE COMPLEXED WITH THE INHIBITOR ZENARESTAT. | | Descriptor: | ALDOSE REDUCTASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, [3-(4-BROMO-2-FLUORO-BENZYL)-7-CHLORO-2,4-DIOXO-3,4-DIHYDRO-2H-QUINAZOLIN-1-YL]-ACETIC ACID | | Authors: | Kinoshita, T, Miyake, H, Fujii, T, Takakura, S, Goto, T. | | Deposit date: | 2001-04-09 | | Release date: | 2002-04-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structure of human recombinant aldose reductase complexed with the potent inhibitor zenarestat.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1J11
 
 | | beta-amylase from Bacillus cereus var. mycoides in complex with alpha-EPG | | Descriptor: | (2R)-oxiran-2-ylmethyl alpha-D-glucopyranoside, Beta-amylase, CALCIUM ION | | Authors: | Oyama, T, Miyake, H, Kusunoki, M, Nitta, Y. | | Deposit date: | 2002-11-25 | | Release date: | 2003-06-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of beta-Amylase from Bacillus cereus var. mycoides in Complexes with Substrate Analogs and Affinity-Labeling Reagents
J.BIOCHEM.(TOKYO), 133, 2003
|
|
1J12
 
 | | Beta-Amylase from Bacillus cereus var. mycoides in Complex with alpha-EBG | | Descriptor: | 2-[(2S)-oxiran-2-yl]ethyl alpha-D-glucopyranoside, Beta-amylase, CALCIUM ION | | Authors: | Oyama, T, Miyake, H, Kusunoki, M, Nitta, Y. | | Deposit date: | 2002-11-25 | | Release date: | 2003-06-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of beta-Amylase from Bacillus cereus var. mycoides in Complexes with Substrate Analogs and Affinity-Labeling Reagents
J.BIOCHEM.(TOKYO), 133, 2003
|
|
1J0Z
 
 | | Beta-amylase from Bacillus cereus var. mycoides in complex with maltose | | Descriptor: | Beta-amylase, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Oyama, T, Miyake, H, Kusunoki, M, Nitta, Y. | | Deposit date: | 2002-11-25 | | Release date: | 2003-06-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structures of beta-Amylase from Bacillus cereus var. mycoides in Complexes with Substrate Analogs and Affinity-Labeling Reagents
J.BIOCHEM.(TOKYO), 133, 2003
|
|
1J0Y
 
 | | Beta-amylase from Bacillus cereus var. mycoides in complex with glucose | | Descriptor: | Beta-amylase, CALCIUM ION, beta-D-glucopyranose | | Authors: | Oyama, T, Miyake, H, Kusunoki, M, Nitta, Y. | | Deposit date: | 2002-11-25 | | Release date: | 2003-06-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of beta-Amylase from Bacillus cereus var. mycoides in Complexes with Substrate Analogs and Affinity-Labeling Reagents
J.BIOCHEM.(TOKYO), 133, 2003
|
|
1TVO
 
 | | The structure of ERK2 in complex with a small molecule inhibitor | | Descriptor: | 5-(2-PHENYLPYRAZOLO[1,5-A]PYRIDIN-3-YL)-1H-PYRAZOLO[3,4-C]PYRIDAZIN-3-AMINE, Mitogen-activated protein kinase 1 | | Authors: | Kinoshita, T. | | Deposit date: | 2004-06-30 | | Release date: | 2005-09-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Identification of a selective ERK inhibitor and structural determination of the inhibitor-ERK2 complex
Biochem.Biophys.Res.Commun., 336, 2005
|
|
1UK1
 
 | |
5TT7
 
 | | Discovery of TAK-659, an Orally Available Investigational Inhibitor of Spleen Tyrosine Kinase (SYK) | | Descriptor: | 2-{[(1R,2S)-2-aminocyclohexyl]amino}-4-[(3-methylphenyl)amino]-6,7-dihydro-5H-pyrrolo[3,4-d]pyrimidin-5-one, Tyrosine-protein kinase SYK | | Authors: | Yano, J, Jennings, A, Lam, B, Hoffman, I.D. | | Deposit date: | 2016-11-01 | | Release date: | 2016-11-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Discovery of TAK-659 an orally available investigational inhibitor of Spleen Tyrosine Kinase (SYK).
Bioorg. Med. Chem. Lett., 26, 2016
|
|
5TR6
 
 | | Discovery of TAK-659, an Orally Available Investigational Inhibitor of Spleen Tyrosine Kinase (SYK) | | Descriptor: | 1,2-ETHANEDIOL, 6-{[(1R,2S)-2-aminocyclohexyl]amino}-7-fluoro-4-(1-methyl-1H-pyrazol-4-yl)-1,2-dihydro-3H-pyrrolo[3,4-c]pyridin-3-one, Tyrosine-protein kinase SYK | | Authors: | Yano, J, Jennings, A, Lam, B, Hoffman, I.D. | | Deposit date: | 2016-10-25 | | Release date: | 2016-11-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Discovery of TAK-659 an orally available investigational inhibitor of Spleen Tyrosine Kinase (SYK).
Bioorg. Med. Chem. Lett., 26, 2016
|
|
5Z5E
 
 | |
3W55
 
 | | The structure of ERK2 in complex with FR148083 | | Descriptor: | (3S,5Z,8S,9S,11E)-8,9,16-trihydroxy-14-methoxy-3-methyl-3,4,9,10-tetrahydro-1H-2-benzoxacyclotetradecine-1,7(8H)-dione, Mitogen-activated protein kinase 1 | | Authors: | Ohori, M, Kinoshita, T. | | Deposit date: | 2013-01-21 | | Release date: | 2013-02-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Role of a cysteine residue in the active site of ERK and the MAPKK family
Biochem.Biophys.Res.Commun., 353, 2007
|
|
