6NRL
 
 | |
6O01
 
 | |
6OQE
 
 | |
7L33
 
 | | X-ray Structure of a Cu-Bound De Novo Designed Peptide Trimer | | Descriptor: | COPPER (II) ION, Cu-3SCC | | Authors: | Chakraborty, S, Wawrzak, Z, Prasad, P, Mitra, S, Prakash, D. | | Deposit date: | 2020-12-17 | | Release date: | 2021-08-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | De Novo Design of a Self-Assembled Artificial Copper Peptide that Activates and Reduces Peroxide
Acs Catalysis, 11, 2021
|
|
1DE9
 
 | | HUMAN APE1 ENDONUCLEASE WITH BOUND ABASIC DNA AND MN2+ ION | | Descriptor: | 5'-d(*CP*TP*AP*C)-3', 5'-d(*GP*AP*TP*CP*GP*GP*TP*AP*G)-3', 5'-d(P*(3DR)P*GP*AP*TP*C)-3', ... | | Authors: | Mol, C.D, Izumi, T, Mitra, S, Tainer, J.A. | | Deposit date: | 1999-11-13 | | Release date: | 2000-02-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | DNA-bound structures and mutants reveal abasic DNA binding by APE1 and DNA repair coordination
Nature, 403, 2000
|
|
1DEW
 
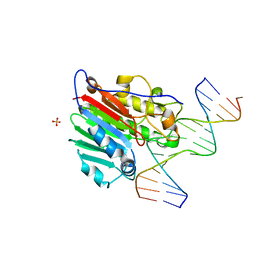 | | CRYSTAL STRUCTURE OF HUMAN APE1 BOUND TO ABASIC DNA | | Descriptor: | 5'-D(*GP*CP*GP*TP*CP*CP*(3DR)P*CP*GP*AP*CP*GP*AP*CP*G)-3', 5'-D(*GP*TP*CP*GP*TP*CP*GP*GP*GP*GP*AP*CP*GP*C)-3', MAJOR APURINIC/APYRIMIDINIC ENDONUCLEASE, ... | | Authors: | Mol, C.D, Izumi, T, Mitra, S, Tainer, J.A. | | Deposit date: | 1999-11-15 | | Release date: | 2000-02-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | DNA-bound structures and mutants reveal abasic DNA binding by APE1 and DNA repair coordination
Nature, 403, 2000
|
|
7ZMZ
 
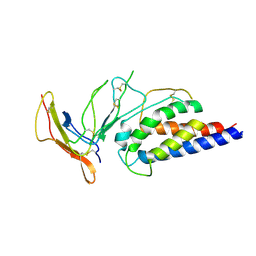 | | Engineered Interleukin 2 bound to CD25 receptor | | Descriptor: | Interleukin-2, Interleukin-2 receptor subunit alpha | | Authors: | Fyfe, P.K, Moraga, I, Gaggero, S, Mitra, S. | | Deposit date: | 2022-04-20 | | Release date: | 2022-12-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | IL-2 is inactivated by the acidic pH environment of tumors enabling engineering of a pH-selective mutein.
Sci Immunol, 7, 2022
|
|
1DE8
 
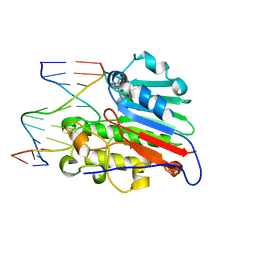 | | HUMAN APURINIC/APYRIMIDINIC ENDONUCLEASE-1 (APE1) BOUND TO ABASIC DNA | | Descriptor: | DNA (5'-D(*CP*GP*AP*TP*CP*GP*GP*TP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*AP*CP*(3DR)P*GP*AP*TP*CP*G)-3'), MAJOR APURINIC/APYRIMIDINIC ENDONUCLEASE | | Authors: | Mol, C.D, Izumi, T, Mitra, S, Tainer, J.A. | | Deposit date: | 1999-11-13 | | Release date: | 2000-02-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | DNA-bound structures and mutants reveal abasic DNA binding by APE1 and DNA repair coordination [corrected
Nature, 403, 2000
|
|
1N2W
 
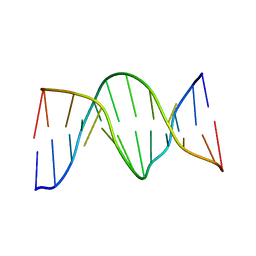 | | Solution Structure of 8OG:G mismatch containing duplex | | Descriptor: | 5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*(8OG)P*GP*CP*G)-3' | | Authors: | Thiviyanathan, V, Somasunderam, A.D, Hazra, T.K, Mitra, S, Gorenstein, D.G. | | Deposit date: | 2002-10-24 | | Release date: | 2002-11-13 | | Last modified: | 2020-02-05 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a DNA Duplex Containing 8-Hydroxy-2'-Deoxyguanosine Opposite Deoxyguanosine
J.Mol.Biol., 325, 2003
|
|
4S1G
 
 | |
2GIU
 
 | |
4XX4
 
 | |
4XX3
 
 | |
4DCQ
 
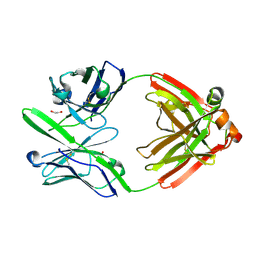 | | Crystal Structure of the Fab Fragment of 3B5H10, an Antibody-Specific for Extended Polyglutamine Repeats (orthorhombic form) | | Descriptor: | 1,2-ETHANEDIOL, 3B5H10 FAB Heavy Chain, 3B5H10 FAB Light Chain | | Authors: | Peters-Libeu, C.A, Tran, T, Finkbeiner, S, Weisgraber, K. | | Deposit date: | 2012-01-18 | | Release date: | 2012-02-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Disease-associated polyglutamine stretches in monomeric huntingtin adopt a compact structure.
J.Mol.Biol., 421, 2012
|
|
5YJJ
 
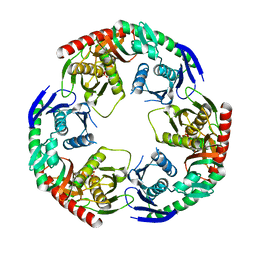 | | Crystal structure of PNPase from Staphylococcus epidermidis | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, Polyribonucleotide nucleotidyltransferase | | Authors: | Raj, R, Gopal, B. | | Deposit date: | 2017-10-10 | | Release date: | 2018-01-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Characterization of Staphylococcus epidermidis Polynucleotide phosphorylase and its interactions with ribonucleases RNase J1 and RNase J2.
Biochem. Biophys. Res. Commun., 495, 2018
|
|
4XHM
 
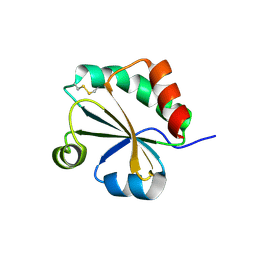 | |
5IWS
 
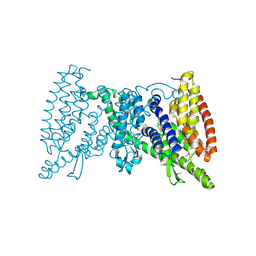 | | Crystal structure of the transporter MalT, the EIIC domain from the maltose-specific phosphotransferase system | | Descriptor: | Protein-N(Pi)-phosphohistidine-sugar phosphotransferase (Enzyme II of the phosphotransferase system) (PTS system glucose-specific IIBC component), alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | McCoy, J.G, Ren, Z, Levin, E.J, Zhou, M, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2016-03-22 | | Release date: | 2016-05-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.551 Å) | | Cite: | The Structure of a Sugar Transporter of the Glucose EIIC Superfamily Provides Insight into the Elevator Mechanism of Membrane Transport.
Structure, 24, 2016
|
|
4GS7
 
 | | Structure of the Interleukin-15 quaternary complex | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ring, A.M, Ozkan, E, Feng, D, Garcia, K.C. | | Deposit date: | 2012-08-27 | | Release date: | 2012-11-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Mechanistic and structural insight into the functional dichotomy between IL-2 and IL-15.
Nat.Immunol., 13, 2012
|
|
4HNO
 
 | | High resolution crystal structure of DNA Apurinic/apyrimidinic (AP) endonuclease IV Nfo from Thermatoga maritima | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MAGNESIUM ION, ... | | Authors: | Shin, D.S, Hosfield, D.J, Arvai, A.S, Tsutakawa, S.E, Tainer, J.A. | | Deposit date: | 2012-10-20 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (0.9194 Å) | | Cite: | Conserved Structural Chemistry for Incision Activity in Structurally Non-homologous Apurinic/Apyrimidinic Endonuclease APE1 and Endonuclease IV DNA Repair Enzymes.
J.Biol.Chem., 288, 2013
|
|
4IEM
 
 | | Human apurinic/apyrimidinic endonuclease (APE1) with product DNA and Mg2+ | | Descriptor: | DNA (5'-D(*CP*GP*AP*TP*CP*GP*GP*TP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*AP*C)-3'), DNA (5'-D(P*(3DR)P*GP*AP*TP*CP*G)-3'), ... | | Authors: | Tsutakawa, S.E, Mol, C.D, Arvai, A.S, Tainer, J.A. | | Deposit date: | 2012-12-13 | | Release date: | 2013-01-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3936 Å) | | Cite: | Conserved Structural Chemistry for Incision Activity in Structurally Non-homologous Apurinic/Apyrimidinic Endonuclease APE1 and Endonuclease IV DNA Repair Enzymes.
J.Biol.Chem., 288, 2013
|
|
3S96
 
 | | Crystal structure of 3B5H10 | | Descriptor: | 3B5H10 FAB heavy chain, 3B5H10 FAB light chain | | Authors: | Weisgraber, K, Peters-Libeu, C, Rutenber, E, Newhouse, Y, Finkbeiner, S. | | Deposit date: | 2011-05-31 | | Release date: | 2012-02-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Disease-associated polyglutamine stretches in monomeric huntingtin adopt a compact structure.
J.Mol.Biol., 421, 2012
|
|
2N8M
 
 | |
2N8L
 
 | |
