1FHM
 
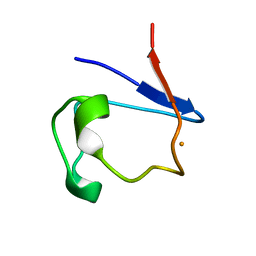 | | X-RAY CRYSTAL STRUCTURE OF REDUCED RUBREDOXIN | | Descriptor: | FE (II) ION, RUBREDOXIN | | Authors: | Min, T, Ergenekan, C.E, Eidsness, M.K, Ichiye, T, Kang, C. | | Deposit date: | 2000-08-02 | | Release date: | 2001-03-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Leucine 41 is a gate for water entry in the reduction of Clostridium pasteurianum rubredoxin.
Protein Sci., 10, 2001
|
|
1FHH
 
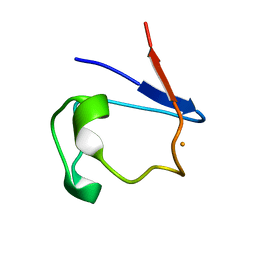 | | X-RAY CRYSTAL STRUCTURE OF OXIDIZED RUBREDOXIN | | Descriptor: | FE (III) ION, RUBREDOXIN | | Authors: | Min, T, Ergenekan, C.E, Eidsness, M.K, Ichiye, T, Kang, C. | | Deposit date: | 2000-08-01 | | Release date: | 2001-03-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Leucine 41 is a gate for water entry in the reduction of Clostridium pasteurianum rubredoxin.
Protein Sci., 10, 2001
|
|
1TSJ
 
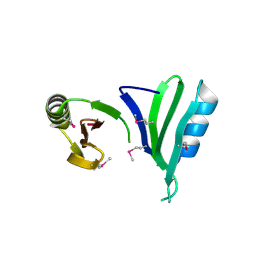 | |
1U6L
 
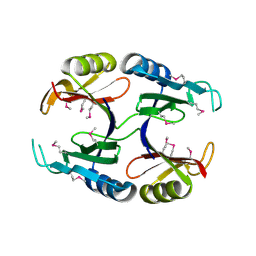 | |
1U6M
 
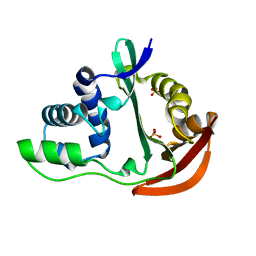 | | The crystal structure of acetyltransferase | | Descriptor: | SULFATE ION, acetyltransferase, GNAT family | | Authors: | Min, T, Gorman, J, Shapiro, L, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-07-30 | | Release date: | 2004-12-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of acetyltransferase, GNAT family from Enterococcus faecalis
To be Published
|
|
1Y9E
 
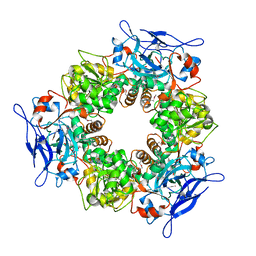 | |
1Y7E
 
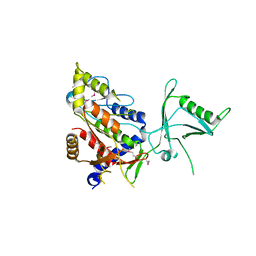 | |
1QYC
 
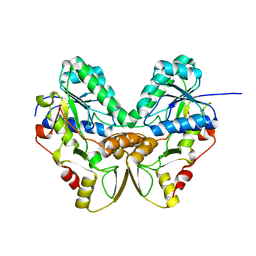 | | Crystal structures of pinoresinol-lariciresinol and phenylcoumaran benzylic ether reductases, and their relationship to isoflavone reductases | | Descriptor: | phenylcoumaran benzylic ether reductase PT1 | | Authors: | Min, T, Kasahara, H, Bedgar, D.L, Youn, B, Lawrence, P.K, Gang, D.R, Halls, S.C, Park, H, Hilsenbeck, J.L, Davin, L.B, Kang, C. | | Deposit date: | 2003-09-10 | | Release date: | 2003-11-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of pinoresinol-lariciresinol and phenylcoumaran benzylic ether reductases and their relationship to isoflavone reductases.
J.Biol.Chem., 278, 2003
|
|
1QYD
 
 | | Crystal structures of pinoresinol-lariciresinol and phenylcoumaran benzylic ether reductases, and their relationship to isoflavone reductases | | Descriptor: | pinoresinol-lariciresinol reductase | | Authors: | Min, T, Kasahara, H, Bedgar, D.L, Youn, B, Lawrence, P.K, Gang, D.R, Halls, S.C, Park, H, Hilsenbeck, J.L, Davin, L.B, Kang, C. | | Deposit date: | 2003-09-10 | | Release date: | 2003-11-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of pinoresinol-lariciresinol and phenylcoumaran benzylic ether reductases and their relationship to isoflavone reductases.
J.Biol.Chem., 278, 2003
|
|
1TT7
 
 | |
2GLJ
 
 | |
2GLF
 
 | |
2I5G
 
 | | Crystal strcuture of amidohydrolase from Pseudomonas aeruginosa | | Descriptor: | amidohydrolase | | Authors: | Min, T, Sauder, J.M, Wasserman, S.R, Smith, D, Burley, S.K, Shapiro, L, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-08-24 | | Release date: | 2006-09-05 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of amidohydrolase from Pseudomonas aeruginosa
To be Published
|
|
2IJZ
 
 | |
3FJ7
 
 | | Crystal structure of L-phospholactate Bound PEB3 | | Descriptor: | L-PHOSPHOLACTATE, Major antigenic peptide PEB3 | | Authors: | Min, T, Matte, A, Cygler, M. | | Deposit date: | 2008-12-14 | | Release date: | 2009-03-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Specificity of Campylobacter jejuni adhesin PEB3 for phosphates and structural differences among its ligand complexes.
Biochemistry, 48, 2009
|
|
3FJG
 
 | | Crystal structure of 3PG bound PEB3 | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, Major antigenic peptide PEB3 | | Authors: | Min, T, Matte, A, Cygler, M. | | Deposit date: | 2008-12-14 | | Release date: | 2009-03-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Specificity of Campylobacter jejuni adhesin PEB3 for phosphates and structural differences among its ligand complexes.
Biochemistry, 48, 2009
|
|
3FIR
 
 | | Crystal structure of Glycosylated K135E PEB3 | | Descriptor: | 2-acetamido-2-deoxy-alpha-L-glucopyranose-(1-3)-2,4-bisacetamido-2,4,6-trideoxy-beta-D-glucopyranose, CITRATE ANION, Major antigenic peptide PEB3 | | Authors: | Min, T, Matte, A, Cygler, M. | | Deposit date: | 2008-12-12 | | Release date: | 2009-03-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Specificity of Campylobacter jejuni adhesin PEB3 for phosphates and structural differences among its ligand complexes.
Biochemistry, 48, 2009
|
|
3FJM
 
 | | crystal structure of phosphate bound PEB3 | | Descriptor: | Major antigenic peptide PEB3, PHOSPHATE ION | | Authors: | Min, T, Matte, A, Cygler, M. | | Deposit date: | 2008-12-14 | | Release date: | 2009-03-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Specificity of Campylobacter jejuni adhesin PEB3 for phosphates and structural differences among its ligand complexes.
Biochemistry, 48, 2009
|
|
1C09
 
 | | RUBREDOXIN V44A CP | | Descriptor: | FE (III) ION, RUBREDOXIN | | Authors: | Min, T, Beard, B, Kang, C. | | Deposit date: | 1999-07-15 | | Release date: | 2001-02-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Modulation of the redox potential of the [Fe(SCys)(4)] site in rubredoxin by the orientation of a peptide dipole.
Biochemistry, 38, 1999
|
|
1CN0
 
 | | CRYSTAL STRUCTURE OF D(ACCCT) | | Descriptor: | DNA (5'-D(*AP*CP*CP*CP*T)-3') | | Authors: | Weil, J, Min, T, Cheng, Y, Wang, S, Sutherland, C, Sinha, N, Kang, C. | | Deposit date: | 1999-05-24 | | Release date: | 2000-05-24 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Stabilization of the i-motif by intramolecular adenine-adenine-thymine base triple in the structure of d(ACCCT).
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1BQJ
 
 | | CRYSTAL STRUCTURE OF D(ACCCT) | | Descriptor: | DNA (5'-D(*AP*CP*CP*CP*T)-3') | | Authors: | Weil, J, Min, T, Yang, C, Wang, S, Sutherland, C, Sinha, N, Kang, C.H. | | Deposit date: | 1998-08-17 | | Release date: | 1999-03-18 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Stabilization of the i-motif by intramolecular adenine-adenine-thymine base triple in the structure of d(ACCCT).
Acta Crystallogr.,Sect.D, 55, 1999
|
|
2NYV
 
 | | X-ray crystal structure of a phosphoglycolate phosphatase from Aquifex aeolicus | | Descriptor: | Phosphoglycolate phosphatase | | Authors: | Ciatto, C, Min, T, Gorman, J, Burley, S.K, Shapiro, L, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-11-21 | | Release date: | 2006-12-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | X-ray crystal structure of a phosphoglycolate phosphatase from Aquifex aeolicus
To be Published
|
|
2OCE
 
 | |
2IJR
 
 | | Crystal structure of a protein api92 from Yersinia pseudotuberculosis, Pfam DUF1281 | | Descriptor: | Hypothetical protein api92 | | Authors: | Jin, X, Min, T, Bonanno, J.B, Sauder, J.M, Wasserman, S, Smith, D, Burley, S.K, Shapiro, L, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-09-30 | | Release date: | 2006-10-31 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of a hypothetical protein from Yersinia
pseudotuberculosis
To be Published
|
|
3C6F
 
 | | Crystal structure of protein Bsu07140 from Bacillus subtilis | | Descriptor: | GLYCEROL, YetF protein | | Authors: | Patskovsky, Y, Min, T, Zhang, A, Adams, J, Groshong, C, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-02-04 | | Release date: | 2008-02-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of protein Bsu07140 from Bacillus subtilis.
To be Published
|
|
