6G16
 
 | | Structure of the human RBBP4:MTA1(464-546) complex showing loop exchange | | Descriptor: | Histone-binding protein RBBP4, Metastasis-associated protein MTA1 | | Authors: | Millard, C.J, Varma, N, Fairall, L, Schwabe, J.W.R. | | Deposit date: | 2018-03-20 | | Release date: | 2018-06-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structure of the core NuRD repression complex provides insights into its interaction with chromatin.
Elife, 5, 2016
|
|
5FXY
 
 | | Structure of the human RBBP4:MTA1(464-546) complex | | Descriptor: | HISTONE-BINDING PROTEIN RBBP4, METASTASIS-ASSOCIATED PROTEIN MTA1 | | Authors: | Millard, C.J, Varma, N, Fairall, L, Schwabe, J.W.R. | | Deposit date: | 2016-03-03 | | Release date: | 2016-05-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The structure of the core NuRD repression complex provides insights into its interaction with chromatin.
Elife, 5, 2016
|
|
7AOA
 
 | | Structure of the extended MTA1/HDAC1/MBD2/RBBP4 NURD deacetylase complex | | Descriptor: | Histone deacetylase 1, Histone-binding protein RBBP4, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Millard, C.J, Fairall, L, Ragan, T.J, Savva, C.G, Schwabe, J.W.R. | | Deposit date: | 2020-10-14 | | Release date: | 2020-11-11 | | Last modified: | 2020-12-30 | | Method: | ELECTRON MICROSCOPY (19.4 Å) | | Cite: | The topology of chromatin-binding domains in the NuRD deacetylase complex.
Nucleic Acids Res., 48, 2020
|
|
7AO9
 
 | | Structure of the core MTA1/HDAC1/MBD2 NURD deacetylase complex | | Descriptor: | Histone deacetylase 1, INOSITOL HEXAKISPHOSPHATE, Metastasis-associated protein MTA1, ... | | Authors: | Millard, C.J, Fairall, L, Ragan, T.J, Savva, C.G, Schwabe, J.W.R. | | Deposit date: | 2020-10-14 | | Release date: | 2020-11-11 | | Last modified: | 2020-12-30 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | The topology of chromatin-binding domains in the NuRD deacetylase complex.
Nucleic Acids Res., 48, 2020
|
|
7AO8
 
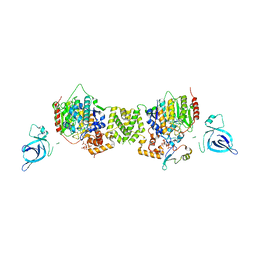 | | Structure of the MTA1/HDAC1/MBD2 NURD deacetylase complex | | Descriptor: | Histone deacetylase 1, INOSITOL HEXAKISPHOSPHATE, Metastasis-associated protein MTA1, ... | | Authors: | Millard, C.J, Fairall, L, Ragan, T.J, Savva, C.G, Schwabe, J.W.R. | | Deposit date: | 2020-10-14 | | Release date: | 2020-11-11 | | Last modified: | 2020-12-30 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | The topology of chromatin-binding domains in the NuRD deacetylase complex.
Nucleic Acids Res., 48, 2020
|
|
2MPM
 
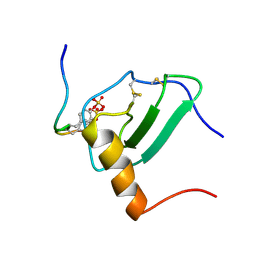 | | Structural Basis of Receptor Sulfotyrosine Recognition by a CC Chemokine: the N-terminal Region of CCR3 Bound to CCL11/Eotaxin-1 | | Descriptor: | CCR3, Eotaxin | | Authors: | Millard, C.J, Ludeman, J.P, Canals, M, Bridgford, J.L, Hinds, M.G, Clayton, D.J, Christopoulos, A, Payne, R.J, Stone, M.J. | | Deposit date: | 2014-05-26 | | Release date: | 2014-12-10 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of Receptor Sulfotyrosine Recognition by a CC Chemokine: The N-Terminal Region of CCR3 Bound to CCL11/Eotaxin-1.
Structure, 22, 2014
|
|
4BKX
 
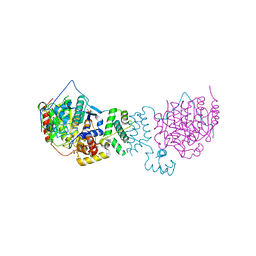 | | The structure of HDAC1 in complex with the dimeric ELM2-SANT domain of MTA1 from the NuRD complex | | Descriptor: | ACETATE ION, HISTONE DEACETYLASE 1, METASTASIS-ASSOCIATED PROTEIN MTA1, ... | | Authors: | Millard, C.J, Watson, P.J, Celardo, I, Gordiyenko, Y, Cowley, S.M, Robinson, C.V, Fairall, L, Schwabe, J.W.R. | | Deposit date: | 2013-04-30 | | Release date: | 2013-07-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Class I Hdacs Share a Common Mechanism of Regulation by Inositol Phosphates.
Mol.Cell, 51, 2013
|
|
5ICN
 
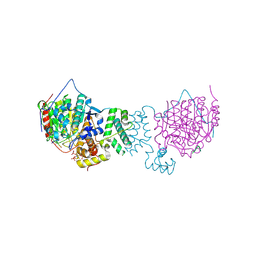 | | HDAC1:MTA1 in complex with inositol-6-phosphate and a novel peptide inhibitor based on histone H4 | | Descriptor: | GLY-ALA-6A0-ARG-HIS, Histone deacetylase 1, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Millard, C.J, Robertson, N.S, Watson, P.J, Jameson, A.G, Schwabe, J.W.R. | | Deposit date: | 2016-02-23 | | Release date: | 2016-05-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Insights into the activation mechanism of class I HDAC complexes by inositol phosphates.
Nat Commun, 7, 2016
|
|
6Z2J
 
 | | The structure of the dimeric HDAC1/MIDEAS/DNTTIP1 MiDAC deacetylase complex | | Descriptor: | Deoxynucleotidyltransferase terminal-interacting protein 1, Histone deacetylase 1, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Fairall, L, Saleh, A, Ragan, T.J, Millard, C.J, Savva, C.G, Schwabe, J.W.R. | | Deposit date: | 2020-05-16 | | Release date: | 2020-07-08 | | Last modified: | 2020-10-07 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | The MiDAC histone deacetylase complex is essential for embryonic development and has a unique multivalent structure.
Nat Commun, 11, 2020
|
|
6Z2K
 
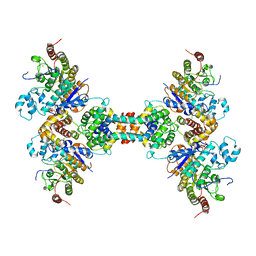 | | The structure of the tetrameric HDAC1/MIDEAS/DNTTIP1 MiDAC deacetylase complex | | Descriptor: | Deoxynucleotidyltransferase terminal-interacting protein 1, Histone deacetylase 1, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Fairall, L, Saleh, A, Ragan, T.J, Millard, C.J, Savva, C.G, Schwabe, J.W.R. | | Deposit date: | 2020-05-16 | | Release date: | 2020-07-08 | | Last modified: | 2020-10-07 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | The MiDAC histone deacetylase complex is essential for embryonic development and has a unique multivalent structure.
Nat Commun, 11, 2020
|
|
2YHN
 
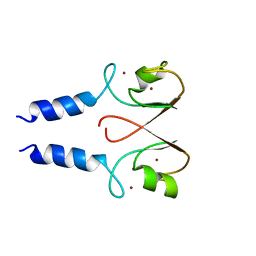 | | The IDOL-UBE2D complex mediates sterol-dependent degradation of the LDL receptor | | Descriptor: | E3 UBIQUITIN-PROTEIN LIGASE MYLIP, ZINC ION | | Authors: | Fairall, L, Goult, B.T, Millard, C.J, Tontonoz, P, Schwabe, J.W.R. | | Deposit date: | 2011-05-04 | | Release date: | 2011-06-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Idol-Ube2D Complex Mediates Sterol-Dependent Degradation of the Ldl Receptor.
Genes Dev., 25, 2011
|
|
2YHO
 
 | | The IDOL-UBE2D complex mediates sterol-dependent degradation of the LDL receptor | | Descriptor: | ACETATE ION, E3 UBIQUITIN-PROTEIN LIGASE MYLIP, UBIQUITIN-CONJUGATING ENZYME E2 D1, ... | | Authors: | Fairall, L, Goult, B.T, Millard, C.J, Tontonoz, P, Schwabe, J.W.R. | | Deposit date: | 2011-05-04 | | Release date: | 2011-06-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Idol-Ube2D Complex Mediates Sterol-Dependent Degradation of the Ldl Receptor.
Genes Dev., 25, 2011
|
|
3EJH
 
 | | Crystal Structure of the Fibronectin 8-9FnI Domain Pair in Complex with a Type-I Collagen Peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Collagen type-I a1 chain, Fibronectin, ... | | Authors: | Erat, M.C, Lowe, E.D, Campbell, I.D, Vakonakis, I. | | Deposit date: | 2008-09-18 | | Release date: | 2009-02-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification and structural analysis of type I collagen sites in complex with fibronectin fragments.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2MWI
 
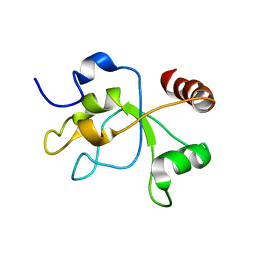 | | The structure of the carboxy-terminal domain of DNTTIP1 | | Descriptor: | Deoxynucleotidyltransferase terminal-interacting protein 1 | | Authors: | Schwabe, J.W.R, Muskett, F.W, Itoh, T. | | Deposit date: | 2014-11-11 | | Release date: | 2015-02-18 | | Last modified: | 2015-03-18 | | Method: | SOLUTION NMR | | Cite: | Structural and functional characterization of a cell cycle associated HDAC1/2 complex reveals the structural basis for complex assembly and nucleosome targeting.
Nucleic Acids Res., 43, 2015
|
|
4D6K
 
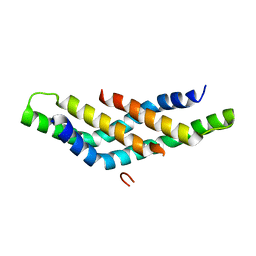 | | Structure of DNTTIP1 dimerisation domain. | | Descriptor: | DEOXYNUCLEOTIDYLTRANSFERASE TERMINAL-INTERACTING PROTEIN 1 | | Authors: | Itoh, T, Fairall, L, Schwabe, J.W.R. | | Deposit date: | 2014-11-11 | | Release date: | 2015-02-18 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Functional Characterization of a Cell Cycle Associated Hdac1/2 Complex Reveals the Structural Basis for Complex Assembly and Nucleosome Targeting.
Nucleic Acids Res., 43, 2015
|
|
