6V8R
 
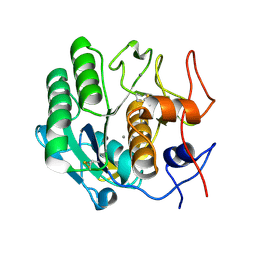 | | Proteinase K Determined by MicroED Phased by ARCIMBOLDO_SHREDDER | | Descriptor: | CALCIUM ION, Proteinase K | | Authors: | Richards, L.S, Martynowycz, M.W, Sawaya, M.R, Millan, C. | | Deposit date: | 2019-12-11 | | Release date: | 2020-08-12 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.6 Å) | | Cite: | Fragment-based determination of a proteinase K structure from MicroED data using ARCIMBOLDO_SHREDDER
Acta Crystallogr.,Sect.D, 76, 2020
|
|
5NDX
 
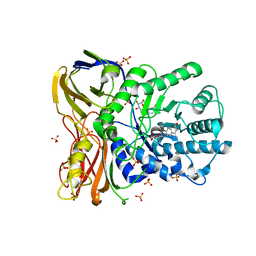 | | The bacterial orthologue of Human a-L-iduronidase does not need N-glycan post-translational modifications to be catalytically competent: Crystallography and QM/MM insights into Mucopolysaccharidosis I | | Descriptor: | (2~{R},3~{S},4~{S},5~{R},6~{S})-6-(4-methyl-2-oxidanylidene-chromen-7-yl)oxy-3,4,5-tris(oxidanyl)oxane-2-carboxylic acid, Glycosyl hydrolase, SULFATE ION | | Authors: | Raich, L, Valero-Gonzalez, J, Castro-Lopez, J, Millan, C, Jimenez-Garcia, M.J, Nieto, P, Uson, I, Hurtado-Guerrero, R, Rovira, C. | | Deposit date: | 2017-03-09 | | Release date: | 2018-07-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The bacterial orthologue of Human a-L-iduronidase does not need N-glycan post-translational modifications to be catalytically competent:
Crystallography and QM/MM insights into Mucopolysaccharidosis I.
To Be Published
|
|
6OJ1
 
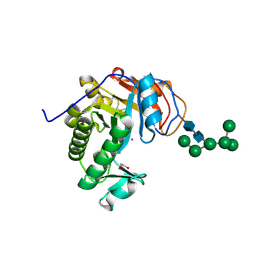 | | Crystal Structure of Aspergillus fumigatus Ega3 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bamford, N.C, Subramanian, A.S, Millan, C, Uson, I, Howell, P.L. | | Deposit date: | 2019-04-10 | | Release date: | 2019-08-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Ega3 from the fungal pathogenAspergillus fumigatusis an endo-alpha-1,4-galactosaminidase that disrupts microbial biofilms.
J.Biol.Chem., 294, 2019
|
|
4LUN
 
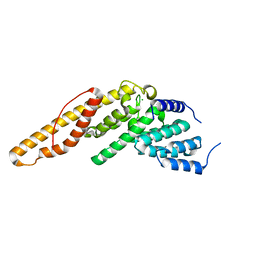 | | Structure of the N-terminal mIF4G domain from S. cerevisiae Upf2, a protein involved in the degradation of mRNAs containing premature stop codons | | Descriptor: | CHLORIDE ION, Nonsense-mediated mRNA decay protein 2 | | Authors: | Fourati, Z, Roy, B, Millan, C, Courreux, P.D, Kervestin, S, van Tilbeurgh, H, He, F, Uson, I, Jacobson, A, Graille, M. | | Deposit date: | 2013-07-25 | | Release date: | 2014-07-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.641 Å) | | Cite: | A highly conserved region essential for NMD in the Upf2 N-terminal domain.
J.Mol.Biol., 426, 2014
|
|
4YT9
 
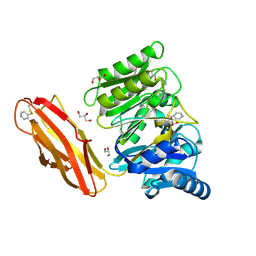 | | Crystal structure of Porphyromonas gingivalis peptidylarginine deiminase (PPAD) substrate-unbound. | | Descriptor: | GLYCEROL, Peptidylarginine deiminase, SODIUM ION | | Authors: | Goulas, T, Mizgalska, D, Garcia-Ferrer, I, Kantyka, T, Guevara, T, Szmigielski, B, Sroka, A, Millan, C, Uson, I, Veillard, F, Potempa, B, Mydel, P, Sola, M, Potempa, J, Gomis-Ruth, F.X. | | Deposit date: | 2015-03-17 | | Release date: | 2015-07-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and mechanism of a bacterial host-protein citrullinating virulence factor, Porphyromonas gingivalis peptidylarginine deiminase.
Sci Rep, 5, 2015
|
|
4YTB
 
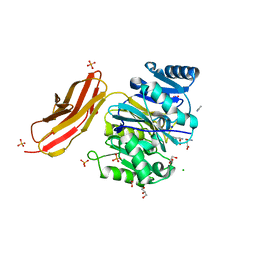 | | Crystal structure of Porphyromonas gingivalis peptidylarginine deiminase (PPAD) in complex with dipeptide Asp-Gln. | | Descriptor: | ASPARTIC ACID, AZIDE ION, CHLORIDE ION, ... | | Authors: | Goulas, T, Mizgalska, D, Garcia-Ferrer, I, Kantyka, T, Guevara, T, Szmigielski, B, Sroka, A, Millan, C, Uson, I, Veillard, F, Potempa, B, Mydel, P, Sola, M, Potempa, J, Gomis-Ruth, F.X. | | Deposit date: | 2015-03-17 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure and mechanism of a bacterial host-protein citrullinating virulence factor, Porphyromonas gingivalis peptidylarginine deiminase.
Sci Rep, 5, 2015
|
|
4YTG
 
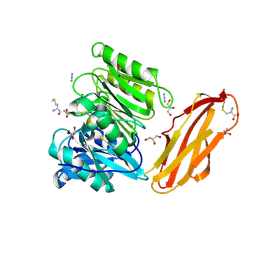 | | Crystal structure of Porphyromonas gingivalis peptidylarginine deiminase (PPAD) mutant C351A in complex with dipeptide Met-Arg. | | Descriptor: | ARGININE, AZIDE ION, CHLORIDE ION, ... | | Authors: | Goulas, T, Mizgalska, D, Garcia-Ferrer, I, Kantyka, T, Guevara, T, Szmigielski, B, Sroka, A, Millan, C, Uson, I, Veillard, F, Potempa, B, Mydel, P, Sola, M, Potempa, J, Gomis-Ruth, F.X. | | Deposit date: | 2015-03-17 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and mechanism of a bacterial host-protein citrullinating virulence factor, Porphyromonas gingivalis peptidylarginine deiminase.
Sci Rep, 5, 2015
|
|
7KFL
 
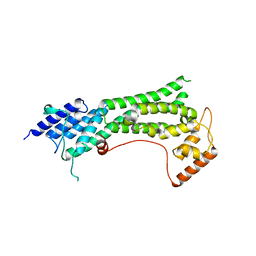 | | Crystal structure of the cargo-binding domain from the plant class XI myosin (MyoXIk) | | Descriptor: | Myosin-17 | | Authors: | Turowski, V.R, Ruiz, D.M, Nascimento, A.F.Z, Millan, C, Sammito, M.D, Juanhuix, J, Cremonesi, A.S, Uson, I, Giuseppe, P.O, Murakami, M.T. | | Deposit date: | 2020-10-14 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of the class XI myosin globular tail reveals evolutionary hallmarks for cargo recognition in plants.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7QEC
 
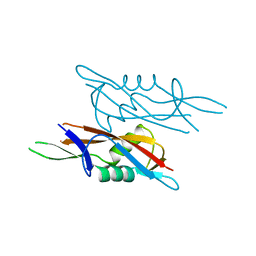 | | Crystal structure of SlpA - domain II, domain that is involved in the self-assembly of the S-layer from Lactobacillus amylovorus | | Descriptor: | S-layer | | Authors: | Eder, M, Dordic, A, Millan, C, Sagmeister, T, Uson, I, Pavkov-Keller, T. | | Deposit date: | 2021-12-02 | | Release date: | 2022-12-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The self-assembly of the S-layer protein from lactobacilli
to be published
|
|
7QFI
 
 | | Crystal structure of S-layer protein SlpX from Lactobacillus acidophilus, domain I (aa 31-182) | | Descriptor: | CALCIUM ION, SlpX | | Authors: | Sagmeister, T, Damisch, E, Millan, C, Uson, I, Eder, M, Pavkov-Keller, T. | | Deposit date: | 2021-12-06 | | Release date: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The self-assembly of the S-layer protein from Lactobacilli acidophilus
To be published
|
|
6F64
 
 | | Crystal structure of the SYCP1 C-terminal back-to-back assembly | | Descriptor: | ACETATE ION, Synaptonemal complex protein 1 | | Authors: | Dunce, J.M, Millan, C, Uson, I, Davies, O.R. | | Deposit date: | 2017-12-04 | | Release date: | 2018-06-06 | | Last modified: | 2020-04-22 | | Method: | X-RAY DIFFRACTION (2.493 Å) | | Cite: | Structural basis of meiotic chromosome synapsis through SYCP1 self-assembly.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6F63
 
 | |
8Q1O
 
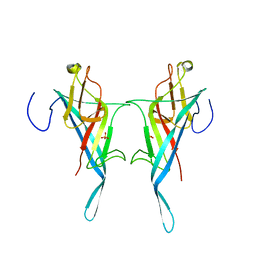 | | S-layer protein SlpA from Lactobacillus amylovorus, domain I (aa 32-209), important for Self-assembly | | Descriptor: | PHOSPHATE ION, S-layer | | Authors: | Sagmeister, T, Grininger, C, Eder, M, Pavkov-Keller, T. | | Deposit date: | 2023-08-01 | | Release date: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.401 Å) | | Cite: | Surface Layer Proteins of Lactobacilli
To Be Published
|
|
4WG0
 
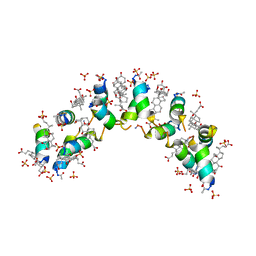 | |
6OJB
 
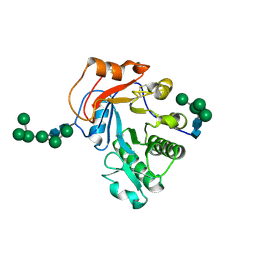 | | Crystal Structure of Aspergillus fumigatus Ega3 complex with galactosamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-amino-2-deoxy-alpha-D-galactopyranose, ... | | Authors: | Bamford, N.C, Howell, P.L. | | Deposit date: | 2019-04-11 | | Release date: | 2019-08-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.093 Å) | | Cite: | Ega3 from the fungal pathogenAspergillus fumigatusis an endo-alpha-1,4-galactosaminidase that disrupts microbial biofilms.
J.Biol.Chem., 294, 2019
|
|
7QFJ
 
 | | Crystal structure of S-layer protein SlpX from Lactobacillus acidophilus, domain II (aa 194-362) | | Descriptor: | SlpX | | Authors: | Sagmeister, T, Pavkov-Keller, T, Buhlheller, C, Baek, M, Read, R, Baker, D. | | Deposit date: | 2021-12-06 | | Release date: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The self-assembly of the S-layer protein from Lactobacilli acidophilus
To be published
|
|
7QFK
 
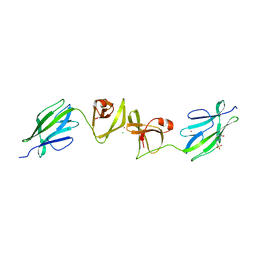 | | Crystal structure of S-layer protein SlpX from Lactobacillus acidophilus, domain II, Co-Crystallization with HgCl2, Mutation Ser316Cys (aa 194-362) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Sagmeister, T, Pavkov-Keller, T, Buhlheller, C. | | Deposit date: | 2021-12-06 | | Release date: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | The self-assembly of the S-layer protein from Lactobacilli acidophilus
To be published
|
|
7QLH
 
 | | Crystal structure of S-layer protein SlpA from Lactobacillus amylovorus, domain I (aa 48-213) | | Descriptor: | PHOSPHATE ION, S-layer, SODIUM ION | | Authors: | Grininger, C, Sagmeister, T, Eder, E, Vejzovic, D, Pavkov-Keller, T. | | Deposit date: | 2021-12-20 | | Release date: | 2022-12-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The self-assembly of the S-layer protein from Lactobacilli acidophilus
to be published
|
|
4GDO
 
 | | Structure of a fragment of the rod domain of plectin | | Descriptor: | Plectin | | Authors: | De Pereda, J.M, Buey, R.M, Uson, I, Sammito, M.D, De Marino, I. | | Deposit date: | 2012-08-01 | | Release date: | 2013-09-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Exploiting tertiary structure through local folds for crystallographic phasing.
Nat.Methods, 10, 2013
|
|
4GN0
 
 | | De novo phasing of a Hamp-complex using an improved Arcimboldo method | | Descriptor: | Hamp domain of AF1503, MAGNESIUM ION | | Authors: | Hulko, M, Ursinus, A, Bar, K, Martin, J, Zeth, K, Lupas, A.N. | | Deposit date: | 2012-08-16 | | Release date: | 2013-09-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Exploiting tertiary structure through local folds for crystallographic phasing.
Nat.Methods, 10, 2013
|
|
7QFL
 
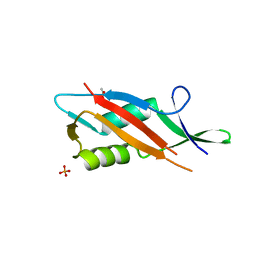 | | Crystal structure of S-layer protein SlpA from Lactobacillus acidophilus, domain II (aa 199-308) | | Descriptor: | ACETATE ION, PHOSPHATE ION, S-layer protein | | Authors: | Sagmeister, T, Dordic, A, Eder, E, Pavkov-Keller, T. | | Deposit date: | 2021-12-06 | | Release date: | 2022-12-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The self-assembly of the S-layer protein from Lactobacilli acidophilus
To be published
|
|
7QLD
 
 | | Crystal structure of S-layer protein SlpA from Lactobacillus acidophilus, domain I, Co-crystallization with HgCl2, Mutation Ser146Cys, (aa 32-198) | | Descriptor: | CHLORIDE ION, MERCURY (II) ION, S-layer protein | | Authors: | Sagmeister, T, Vejzovic, D, Eder, M, Dordic, A, Pavkov-Keller, T. | | Deposit date: | 2021-12-20 | | Release date: | 2022-12-28 | | Method: | X-RAY DIFFRACTION (2.153 Å) | | Cite: | The self-assembly of the S-layer protein from Lactobacilli acidophilus
To Be Published
|
|
7QLE
 
 | | Crystal structure of S-layer protein SlpA from Lactobacillus acidophilus, domain I (aa 32-198) | | Descriptor: | S-layer protein | | Authors: | Sagmeister, T, Eder, M, Vejzovic, D, Dordic, A, Pavkov-Keller, T. | | Deposit date: | 2021-12-20 | | Release date: | 2022-12-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The self-assembly of the S-layer protein from Lactobacilli acidophilus
To Be Published
|
|
7N2H
 
 | | X-Ray structure of a sequence variant of a repeat segment of the yeast prion New1p | | Descriptor: | prion New1p | | Authors: | Richards, L.S, Flores, M.D, Zee, C.T, Glynn, C, Gallagher-Jones, M, Sawaya, M.R. | | Deposit date: | 2021-05-29 | | Release date: | 2022-06-01 | | Last modified: | 2023-05-10 | | Method: | X-RAY DIFFRACTION (1.102 Å) | | Cite: | Fragment-Based Ab Initio Phasing of Peptidic Nanocrystals by MicroED.
Acs Bio Med Chem Au, 3, 2023
|
|
7N2G
 
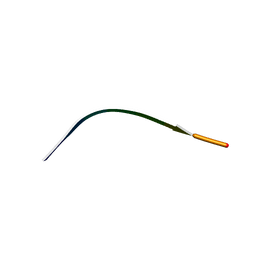 | | MicroED structure of human CPEB3 segment(154-161) kinked polymorph phased by ARCIMBOLDO-BORGES | | Descriptor: | CPEB3 | | Authors: | Flores, M.D, Richards, L.S, Zee, C.T, Glynn, C, Gallagher-Jones, M, Sawaya, M.R. | | Deposit date: | 2021-05-29 | | Release date: | 2022-06-01 | | Last modified: | 2023-05-10 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.201 Å) | | Cite: | Fragment-Based Ab Initio Phasing of Peptidic Nanocrystals by MicroED.
Acs Bio Med Chem Au, 3, 2023
|
|
