2O6L
 
 | |
1PQZ
 
 | |
1RJZ
 
 | | Mhc Class I Natural Mutant H-2Kbm8 Heavy Chain Complexed With beta-2 Microglobulin and Herpies Simplex Virus Mutant Glycoprotein B Peptide | | Descriptor: | Beta-2-microglobulin, Glycoprotein B, H-2 class I histocompatibility antigen, ... | | Authors: | Miley, M.J, Messaoudi, I, Nikolich-Zugich, J, Fremont, D.H. | | Deposit date: | 2003-11-20 | | Release date: | 2004-12-14 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis for the Restoration of TCR Recognition of an MHC Allelic Variant by Peptide Secondary Anchor Substitution
J.Exp.Med., 200, 2004
|
|
1RJY
 
 | | Mhc Class I Natural Mutant H-2Kbm8 Heavy Chain Complexed With beta-2 Microglobulin and Herpes Simplex Virus Glycoprotein B Peptide | | Descriptor: | Beta-2-microglobulin, Glycoprotein B, H-2 class I histocompatibility antigen, ... | | Authors: | Miley, M.J, Messaoudi, I, Nikolich-Zugich, J, Fremont, D.H. | | Deposit date: | 2003-11-20 | | Release date: | 2004-12-14 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for the Restoration of TCR Recognition of an MHC Allelic Variant by Peptide Secondary Anchor Substitution
J.Exp.Med., 200, 2004
|
|
1RK1
 
 | | Mhc Class I Natural H-2Kb Heavy Chain Complexed With beta-2 Microglobulin and Herpes Simplex Virus Mutant Glycoprotein B Peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, Glycoprotein B, ... | | Authors: | Miley, M.J, Messaoudi, I, Nikolich-Zugich, J, Fremont, D.H. | | Deposit date: | 2003-11-20 | | Release date: | 2004-12-14 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for the Restoration of TCR Recognition of an MHC Allelic Variant by Peptide Secondary Anchor Substitution
J.Exp.Med., 200, 2004
|
|
1RK0
 
 | | Mhc Class I H-2Kb Heavy Chain Complexed With beta-2 Microglobulin and Herpes Simplex Virus Glycoprotein B peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, Glycoprotein B, ... | | Authors: | Miley, M.J, Messaoudi, I, Nikolich-Zugich, J, Fremont, D.H. | | Deposit date: | 2003-11-20 | | Release date: | 2004-12-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural Basis for the Restoration of TCR Recognition of an MHC Allelic Variant by Peptide Secondary Anchor Substitution
J.Exp.Med., 200, 2004
|
|
1FNE
 
 | | HISTOCOMPATIBILITY ANTIGEN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEIN (MHC CLASS II I-EK, ALPHA CHAIN), ... | | Authors: | Miley, M.J, Nelson, C.A, Fremont, D.H. | | Deposit date: | 2000-08-21 | | Release date: | 2001-03-07 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and functional consequences of altering a peptide MHC anchor residue.
J.Immunol., 166, 2001
|
|
1FNG
 
 | | HISTOCOMPATIBILITY ANTIGEN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEIN (MHC CLASS II I-EK, ALPHA CHAIN), ... | | Authors: | Miley, M.J, Nelson, C.A, Fremont, D.H. | | Deposit date: | 2000-08-21 | | Release date: | 2001-03-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and functional consequences of altering a peptide MHC anchor residue.
J.Immunol., 166, 2001
|
|
5K0K
 
 | | Crystal structure of the catalytic domain of the proto-oncogene tyrosine-protein kinase MER in complex with inhibitor UNC2434 | | Descriptor: | 15-{4-[(4-methylpiperazin-1-yl)methyl]phenyl}-4,5,6,7,9,10,11,12-octahydro-2,16-(azenometheno)pyrrolo[2,1-d][1,3,5,9]te traazacyclotetradecin-8(3H)-one, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Wang, X, Liu, J, Zhang, W, Stashko, M.A, Nichols, J, DeRyckere, D, Miley, M.J, Norris-Drouin, J, Chen, Z, Machius, M, Wood, E, Graham, D.K, Earp, H.S, Graham, K, Kireev, D, Frye, S.V. | | Deposit date: | 2016-05-17 | | Release date: | 2017-01-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.545 Å) | | Cite: | Design and Synthesis of Novel Macrocyclic Mer Tyrosine Kinase Inhibitors.
ACS Med Chem Lett, 7, 2016
|
|
5K0X
 
 | | Crystal structure of the catalytic domain of the proto-oncogene tyrosine-protein kinase MER in complex with inhibitor UNC2541 | | Descriptor: | (7S)-7-amino-N-[(4-fluorophenyl)methyl]-8-oxo-2,9,16,18,21-pentaazabicyclo[15.3.1]henicosa-1(21),17,19-triene-20-carboxamide, CHLORIDE ION, Tyrosine-protein kinase Mer | | Authors: | McIver, A.L, Zhang, W, Liu, Q, Jiang, X, Stashko, M.A, Nichols, J, Miley, M.J, Norris-Drouin, J, Machius, M, DeRyckere, D, Wood, E, Graham, D.K, Earp, H.S, Kireev, D, Frye, S.V, Wang, X. | | Deposit date: | 2016-05-17 | | Release date: | 2017-02-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.231 Å) | | Cite: | Discovery of Macrocyclic Pyrimidines as MerTK-Specific Inhibitors.
ChemMedChem, 12, 2017
|
|
3ONW
 
 | | Structure of a G-alpha-i1 mutant with enhanced affinity for the RGS14 GoLoco motif. | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Guanine nucleotide-binding protein G(i) subunit alpha-1, Regulator of G-protein signaling 14, ... | | Authors: | Bosch, D, Kimple, A.J, Sammond, D.W, Miley, M.J, Machius, M, Kuhlman, B, Willard, F.S, Siderovski, D.P. | | Deposit date: | 2010-08-30 | | Release date: | 2010-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structural Determinants of Affinity Enhancement between GoLoco Motifs and G-Protein {alpha} Subunit Mutants.
J.Biol.Chem., 286, 2011
|
|
4MH7
 
 | | Crystal structure of the catalytic domain of the proto-oncogene tyrosine-protein kinase MER in complex with inhibitor UNC1896 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, N-butyl-2-(butylamino)-4-[(trans-4-hydroxycyclohexyl)amino]-N-methylpyrimidine-5-carboxamide, ... | | Authors: | Zhang, W, McIver, A, Stashko, M.A, Deryckere, D, Branchford, B.R, Hunter, D, Kireev, D.B, Miley, M.J, Norris-Drouin, J, Stewart, W.M, Lee, M, Sather, S, Zhou, Y, DiPaola, J.A, Machius, M, Janzen, W.P, Earp, H.S, Graham, D.K, Frye, S, Wang, X. | | Deposit date: | 2013-08-29 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Discovery of Mer specific tyrosine kinase inhibitors for the treatment and prevention of thrombosis.
J.Med.Chem., 56, 2013
|
|
3V1A
 
 | | Crystal structure of de novo designed MID1-apo1 | | Descriptor: | Computational design, MID1-apo1 | | Authors: | Der, B.S, Machius, M, Miley, M.J, Kuhlman, B. | | Deposit date: | 2011-12-09 | | Release date: | 2012-01-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Metal-mediated affinity and orientation specificity in a computationally designed protein homodimer.
J.Am.Chem.Soc., 134, 2012
|
|
3V1E
 
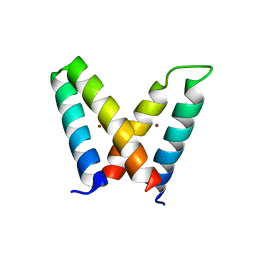 | | Crystal structure of de novo designed MID1-zinc H12E mutant | | Descriptor: | Computational design, MID1-zinc H12E mutant, ZINC ION | | Authors: | Der, B.S, Machius, M, Miley, M.J, Kuhlman, B. | | Deposit date: | 2011-12-09 | | Release date: | 2012-01-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.073 Å) | | Cite: | Metal-mediated affinity and orientation specificity in a computationally designed protein homodimer.
J.Am.Chem.Soc., 134, 2012
|
|
3V1F
 
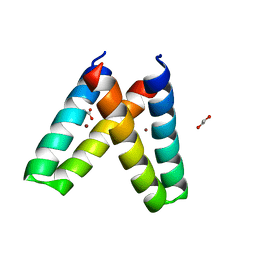 | | Crystal structure of de novo designed MID1-zinc H35E mutant | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Computational design, ... | | Authors: | Der, B.S, Machius, M, Miley, M.J, Kuhlman, B. | | Deposit date: | 2011-12-09 | | Release date: | 2012-01-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.151 Å) | | Cite: | Metal-mediated affinity and orientation specificity in a computationally designed protein homodimer.
J.Am.Chem.Soc., 134, 2012
|
|
3V1C
 
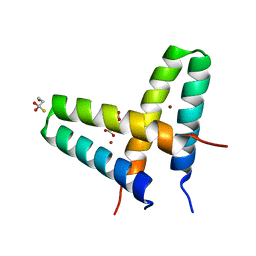 | | Crystal structure of de novo designed MID1-zinc | | Descriptor: | Computational design, MID1-zinc, L(+)-TARTARIC ACID, ... | | Authors: | Der, B.S, Machius, M, Miley, M.J, Kuhlman, B. | | Deposit date: | 2011-12-09 | | Release date: | 2012-01-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.129 Å) | | Cite: | Metal-mediated affinity and orientation specificity in a computationally designed protein homodimer.
J.Am.Chem.Soc., 134, 2012
|
|
3V1D
 
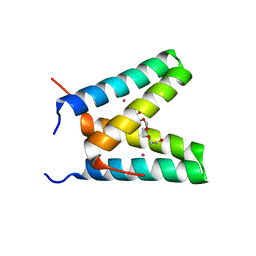 | | Crystal structure of de novo designed MID1-cobalt | | Descriptor: | COBALT (II) ION, Computational design, MID1-cobalt, ... | | Authors: | Der, B.S, Machius, M, Miley, M.J, Kuhlman, B. | | Deposit date: | 2011-12-09 | | Release date: | 2012-01-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.239 Å) | | Cite: | Metal-mediated affinity and orientation specificity in a computationally designed protein homodimer.
J.Am.Chem.Soc., 134, 2012
|
|
3V1B
 
 | | Crystal structure of de novo designed MID1-apo2 | | Descriptor: | Computational design, MID1-apo2, GLYCEROL | | Authors: | Der, B.S, Machius, M, Miley, M.J, Kuhlman, B. | | Deposit date: | 2011-12-09 | | Release date: | 2012-01-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Metal-mediated affinity and orientation specificity in a computationally designed protein homodimer.
J.Am.Chem.Soc., 134, 2012
|
|
3ZY7
 
 | | Crystal structure of computationally redesigned gamma-adaptin appendage domain forming a symmetric homodimer | | Descriptor: | AP-1 COMPLEX SUBUNIT GAMMA-1, DI(HYDROXYETHYL)ETHER, ISOPROPYL ALCOHOL | | Authors: | Stranges, P.B, Machius, M, Miley, M.J, Tripathy, A, Kuhlman, B. | | Deposit date: | 2011-08-17 | | Release date: | 2011-12-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Computational Design of a Symmetric Homodimer Using Beta-Strand Assembly.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
6MEP
 
 | | Crystal structure of the catalytic domain of the proto-oncogene tyrosine-protein kinase MER in complex with inhibitor UNC3437 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Tyrosine-protein kinase Mer, ... | | Authors: | Da, C, Zhang, D, Stashko, M.A, Cheng, A, Hunter, D, Norris-Drouin, J, Graves, L, Machius, M, Miley, M.J, DeRyckere, D, Earp, H.S, Graham, D.K, Frye, S.V, Wang, X, Kireev, D. | | Deposit date: | 2018-09-06 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.893 Å) | | Cite: | Data-Driven Construction of Antitumor Agents with Controlled Polypharmacology.
J.Am.Chem.Soc., 141, 2019
|
|
4M3Q
 
 | | Crystal structure of the catalytic domain of the proto-oncogene tyrosine-protein kinase MER in complex with inhibitor UNC1917 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Tyrosine-protein kinase Mer, ... | | Authors: | Zhang, W, Zhang, D, Stashko, M.A, DeRyckere, D, Hunter, D, Kireev, D.B, Miley, M, Cummings, C, Lee, M, Norris-Drouin, J, Stewart, W.M, Sather, S, Zhou, Y, Kirkpatrick, G, Machius, M, Janzen, W.P, Earp, H.S, Graham, D.K, Frye, S, Wang, X. | | Deposit date: | 2013-08-06 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.718 Å) | | Cite: | Pseudo-Cyclization through Intramolecular Hydrogen Bond Enables Discovery of Pyridine Substituted Pyrimidines as New Mer Kinase Inhibitors.
J.Med.Chem., 56, 2013
|
|
4MHA
 
 | | Crystal structure of the catalytic domain of the proto-oncogene tyrosine-protein kinase MER in complex with inhibitor UNC1817 | | Descriptor: | 2-(butylamino)-4-[(trans-4-hydroxycyclohexyl)amino]-N-(4-sulfamoylbenzyl)pyrimidine-5-carboxamide, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Zhang, W, Mciver, A, Stashko, M.A, Deryckere, D, Branchford, B.R, Hunter, D, Kireev, D.B, Miley, D.B.M, Norris-Drouin, J, Stewart, W.M, Lee, M, Sather, S, Zhou, Y, Dipaola, J.A, Machius, M, Janzen, W.P, Earp, H.S, Graham, D.K, Frye, S, Wang, X. | | Deposit date: | 2013-08-29 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Discovery of Mer specific tyrosine kinase inhibitors for the treatment and prevention of thrombosis.
J.Med.Chem., 56, 2013
|
|
5C7I
 
 | |
5C7O
 
 | | Structure of human testis-specific glyceraldehyde-3-phosphate dehydrogenase holo form with NAD+ | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase, testis-specific, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Betts, L, Machius, M, Danshina, P, O'Brien, D. | | Deposit date: | 2015-06-24 | | Release date: | 2016-03-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.729 Å) | | Cite: | Structural analyses to identify selective inhibitors of glyceraldehyde 3-phosphate dehydrogenase-S, a sperm-specific glycolytic enzyme.
Mol. Hum. Reprod., 22, 2016
|
|
5C7L
 
 | | Structure of human testis-specific glyceraldehyde-3-phosphate dehydrogenase apo form | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase, testis-specific | | Authors: | Betts, L, Machius, M, Danshina, P, O'Brien, D. | | Deposit date: | 2015-06-24 | | Release date: | 2016-03-09 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (1.862 Å) | | Cite: | Structural analyses to identify selective inhibitors of glyceraldehyde 3-phosphate dehydrogenase-S, a sperm-specific glycolytic enzyme.
Mol. Hum. Reprod., 22, 2016
|
|
