6HNJ
 
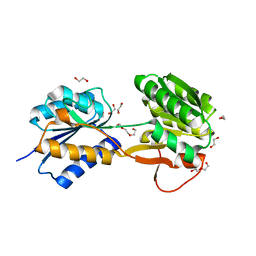 | | The ligand-bound, open structure of CD0873, a substrate binding protein with adhesive properties from Clostridium difficile. | | Descriptor: | 1,2-ETHANEDIOL, ABC-type transport system, sugar-family extracellular solute-binding protein, ... | | Authors: | Bradshaw, W.J, Kovacs-Simon, A, Harmer, N.J, Michell, S.L, Acharya, K.R. | | Deposit date: | 2018-09-15 | | Release date: | 2019-08-28 | | Last modified: | 2019-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular features of lipoprotein CD0873: A potential vaccine against the human pathogenClostridioides difficile.
J.Biol.Chem., 294, 2019
|
|
6HNI
 
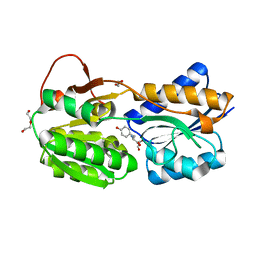 | | The ligand-bound, closed structure of CD0873, a substrate binding protein with adhesive properties from Clostridium difficile. | | Descriptor: | 1,2-ETHANEDIOL, ABC-type transport system, sugar-family extracellular solute-binding protein, ... | | Authors: | Bradshaw, W.J, Kovacs-Simon, A, Harmer, N.J, Michell, S.L, Acharya, K.R. | | Deposit date: | 2018-09-15 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Molecular features of lipoprotein CD0873: A potential vaccine against the human pathogenClostridioides difficile.
J.Biol.Chem., 294, 2019
|
|
6HNK
 
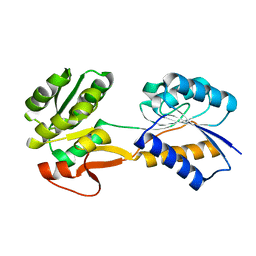 | | The ligand-free, open structure of CD0873, a substrate binding protein with adhesive properties from Clostridium difficile. | | Descriptor: | ABC-type transport system, sugar-family extracellular solute-binding protein | | Authors: | Bradshaw, W.J, Kovacs-Simon, A, Harmer, N.J, Michell, S.L, Acharya, K.R. | | Deposit date: | 2018-09-15 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular features of lipoprotein CD0873: A potential vaccine against the human pathogenClostridioides difficile.
J.Biol.Chem., 294, 2019
|
|
5TKQ
 
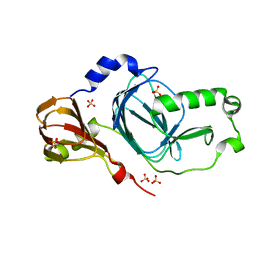 | | Crystal structure of human 3HAO with zinc bound in the active site | | Descriptor: | 3-hydroxyanthranilate 3,4-dioxygenase, SULFATE ION, ZINC ION | | Authors: | Pidugu, L.S, Toth, E.A. | | Deposit date: | 2016-10-07 | | Release date: | 2017-04-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structures of human 3-hydroxyanthranilate 3,4-dioxygenase with native and non-native metals bound in the active site.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5TK5
 
 | | Crystal structure of human 3HAO with iron bound in the active site | | Descriptor: | 3-hydroxyanthranilate 3,4-dioxygenase, FE (III) ION, SULFATE ION | | Authors: | Pidugu, L.S, Toth, E.A. | | Deposit date: | 2016-10-06 | | Release date: | 2017-04-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal structures of human 3-hydroxyanthranilate 3,4-dioxygenase with native and non-native metals bound in the active site.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
3PHT
 
 | |
3QSI
 
 | |
3LGH
 
 | |
4B5C
 
 | | Crystal structure of the peptidoglycan-associated lipoprotein from Burkholderia pseudomallei | | Descriptor: | ACETATE ION, PUTATIVE OMPA FAMILY LIPOPROTEIN | | Authors: | Gourlay, L.J, Peri, C, Conchillo-Sole, O, Ferrer-Navarro, M, Gori, A, Longhi, R, Rinchai, D, Lertmemongkolchai, G, Lassaux, P, Daura, X, Colombo, G, Bolognesi, M. | | Deposit date: | 2012-08-03 | | Release date: | 2013-08-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Exploiting the Burkholderia Pseudomallei Acute Phase Antigen Bpsl2765 for Structure-Based Epitope Discovery/Design in Structural Vaccinology.
Chem.Biool., 20, 2013
|
|
