4W55
 
 | | T4 Lysozyme L99A with n-Propylbenzene Bound | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Endolysin, propylbenzene | | Authors: | Merski, M, Shoichet, B.K, Eidam, O, Fischer, M. | | Deposit date: | 2014-08-16 | | Release date: | 2015-04-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6401 Å) | | Cite: | Homologous ligands accommodated by discrete conformations of a buried cavity.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4W58
 
 | | T4 Lysozyme L99A with n-Pentylbenzene Bound | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Endolysin, pentylbenzene | | Authors: | Merski, M, Shoichet, B.K, Eidam, O, Fischer, M. | | Deposit date: | 2014-08-16 | | Release date: | 2015-04-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Homologous ligands accommodated by discrete conformations of a buried cavity.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4W52
 
 | | T4 Lysozyme L99A with Benzene Bound | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, BENZENE, Endolysin | | Authors: | Merski, M, Shoichet, B.K, Eidam, O, Fischer, M. | | Deposit date: | 2014-08-16 | | Release date: | 2015-04-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5001 Å) | | Cite: | Homologous ligands accommodated by discrete conformations of a buried cavity.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4W54
 
 | | T4 Lysozyme L99A with Ethylbenzene Bound | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Endolysin, PHENYLETHANE | | Authors: | Merski, M, Shoichet, B.K, Eidam, O, Fischer, M. | | Deposit date: | 2014-08-16 | | Release date: | 2015-04-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7901 Å) | | Cite: | Homologous ligands accommodated by discrete conformations of a buried cavity.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4W57
 
 | | T4 Lysozyme L99A with n-Butylbenzene Bound | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Endolysin, N-BUTYLBENZENE | | Authors: | Merski, M, Shoichet, B.K, Eidam, O, Fischer, M. | | Deposit date: | 2014-08-16 | | Release date: | 2015-04-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6801 Å) | | Cite: | Homologous ligands accommodated by discrete conformations of a buried cavity.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4W51
 
 | | T4 Lysozyme L99A with No Ligand Bound | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Endolysin | | Authors: | Merski, M, Shoichet, B.K, Eidam, O, Fischer, M. | | Deposit date: | 2014-08-16 | | Release date: | 2015-04-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Homologous ligands accommodated by discrete conformations of a buried cavity.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4W53
 
 | | T4 Lysozyme L99A with Toluene Bound | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Endolysin, TOLUENE | | Authors: | Merski, M, Shoichet, B.K, Eidam, O, Fischer, M. | | Deposit date: | 2014-08-16 | | Release date: | 2015-04-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Homologous ligands accommodated by discrete conformations of a buried cavity.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4W56
 
 | | T4 Lysozyme L99A with sec-Butylbenzene Bound | | Descriptor: | (2R)-butan-2-ylbenzene, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Endolysin | | Authors: | Merski, M, Shoichet, B.K, Eidam, O, Fischer, M. | | Deposit date: | 2014-08-16 | | Release date: | 2015-04-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Homologous ligands accommodated by discrete conformations of a buried cavity.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4W59
 
 | | T4 Lysozyme L99A with n-Hexylbenzene Bound | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Endolysin, hexylbenzene | | Authors: | Merski, M, Shoichet, B.K, Eidam, O, Fischer, M. | | Deposit date: | 2014-08-16 | | Release date: | 2015-04-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Homologous ligands accommodated by discrete conformations of a buried cavity.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5M3O
 
 | | HTRA2 A141S mutant structure | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Serine protease HTRA2, mitochondrial | | Authors: | Merski, M, Barbosa Pereira, P.J, Macedo-Ribeiro, S. | | Deposit date: | 2016-10-15 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular motion regulates the activity of the Mitochondrial Serine Protease HtrA2.
Cell Death Dis, 8, 2017
|
|
5M3N
 
 | | HTRA2 wild-type structure | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Serine protease HTRA2, mitochondrial | | Authors: | Merski, M, Pereira, P.J.B, Macedo-Ribeiro, S. | | Deposit date: | 2016-10-15 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.649 Å) | | Cite: | Molecular motion regulates the activity of the Mitochondrial Serine Protease HtrA2.
Cell Death Dis, 8, 2017
|
|
5TNY
 
 | | HTRA2 G399S mutant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Serine protease HTRA2, mitochondrial | | Authors: | Merski, M, Barbosa Pereira, P.J, Macedo-Ribeiro, S. | | Deposit date: | 2016-10-15 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular motion regulates the activity of the Mitochondrial Serine Protease HtrA2.
Cell Death Dis, 8, 2017
|
|
5TO0
 
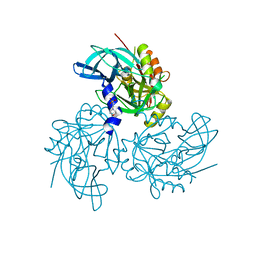 | | HTRA2 S276C mutant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Serine protease HTRA2, mitochondrial | | Authors: | Merski, M, Barbosa Pereira, P.J, Macedo-Ribeiro, S. | | Deposit date: | 2016-10-15 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular motion regulates the activity of the Mitochondrial Serine Protease HtrA2.
Cell Death Dis, 8, 2017
|
|
5TO1
 
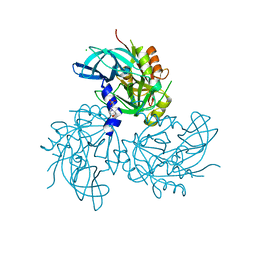 | | HtrA2 exposed (L266R, F303A) mutant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, Serine protease HTRA2, ... | | Authors: | Merski, M, Barbosa Pereira, P.J, Macedo-Ribeiro, S. | | Deposit date: | 2016-10-15 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Molecular motion regulates the activity of the Mitochondrial Serine Protease HtrA2.
Cell Death Dis, 8, 2017
|
|
4EKS
 
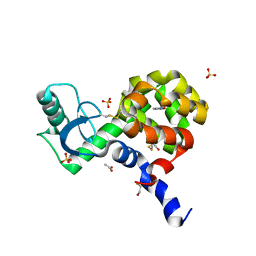 | | T4 Lysozyme L99A/M102H with Isoxazole Bound | | Descriptor: | 1,2-benzisoxazole, 2-HYDROXYETHYL DISULFIDE, ACETATE ION, ... | | Authors: | Merski, M, Shoichet, B.K. | | Deposit date: | 2012-04-09 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Engineering a model protein cavity to catalyze the Kemp elimination.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4E97
 
 | |
4EKP
 
 | | T4 Lysozyme L99A/M102H with Nitrobenzene Bound | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, ACETATE ION, BETA-MERCAPTOETHANOL, ... | | Authors: | Merski, M, Shoichet, B.K. | | Deposit date: | 2012-04-09 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Engineering a model protein cavity to catalyze the Kemp elimination.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4EKQ
 
 | | T4 Lysozyme L99A/M102H with 4-Nitrophenol Bound | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, ACETATE ION, BETA-MERCAPTOETHANOL, ... | | Authors: | Merski, M, Shoichet, B.K. | | Deposit date: | 2012-04-09 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Engineering a model protein cavity to catalyze the Kemp elimination.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4EKR
 
 | | T4 Lysozyme L99A/M102H with 2-Cyanophenol Bound | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, 2-hydroxybenzonitrile, ACETATE ION, ... | | Authors: | Merski, M, Shoichet, B.K. | | Deposit date: | 2012-04-09 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Engineering a model protein cavity to catalyze the Kemp elimination.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4I7R
 
 | |
4I7J
 
 | | T4 Lysozyme L99A/M102H with benzene bound | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, ACETATE ION, BENZENE, ... | | Authors: | Merski, M, Shoichet, B.K. | | Deposit date: | 2012-11-30 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | The impact of introducing a histidine into an apolar cavity site on docking and ligand recognition.
J.Med.Chem., 56, 2013
|
|
4I7T
 
 | |
4I7O
 
 | |
4I7N
 
 | | T4 Lysozyme L99A/M102H with 1-phenyl-2-propyn-1-ol bound | | Descriptor: | (1R)-1-phenylprop-2-yn-1-ol, 2-HYDROXYETHYL DISULFIDE, ACETATE ION, ... | | Authors: | Merski, M, Shoichet, B.K. | | Deposit date: | 2012-11-30 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | The impact of introducing a histidine into an apolar cavity site on docking and ligand recognition.
J.Med.Chem., 56, 2013
|
|
4I7K
 
 | | T4 Lysozyme L99A/M102H with toluene bound | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, ACETATE ION, BETA-MERCAPTOETHANOL, ... | | Authors: | Merski, M, Shoichet, B.K. | | Deposit date: | 2012-11-30 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | The impact of introducing a histidine into an apolar cavity site on docking and ligand recognition.
J.Med.Chem., 56, 2013
|
|
