8WZB
 
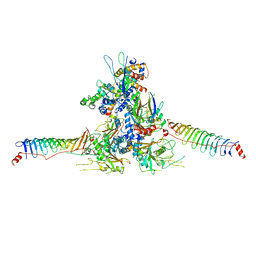 | | RS head-neck monomer | | Descriptor: | DPY30 domain containing 2, DnaJ homolog subfamily B member 13, Nucleoside diphosphate kinase homolog 5, ... | | Authors: | Meng, X, Cong, Y. | | Deposit date: | 2023-11-01 | | Release date: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Multi-scale structures of the mammalian radial spoke and divergence of axonemal complexes in ependymal cilia.
Nat Commun, 15, 2024
|
|
8X2U
 
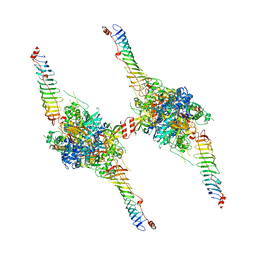 | | Radial spoke head-neck dimer | | Descriptor: | DPY30 domain containing 2, DnaJ homolog subfamily B member 13, Nucleoside diphosphate kinase homolog 5, ... | | Authors: | Meng, X, Cong, Y. | | Deposit date: | 2023-11-10 | | Release date: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Multi-scale structures of the mammalian radial spoke and divergence of axonemal complexes in ependymal cilia.
Nat Commun, 15, 2024
|
|
5CYW
 
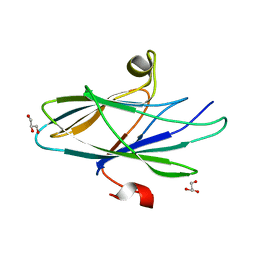 | | Crystal Structure of Vaccinia Virus C7 | | Descriptor: | GLYCEROL, Interferon antagonist C7 | | Authors: | Krumm, B.E, Meng, X, Li, Y, Xiang, Y, Deng, J. | | Deposit date: | 2015-07-30 | | Release date: | 2015-11-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for antagonizing a host restriction factor by C7 family of poxvirus host-range proteins.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5CZ3
 
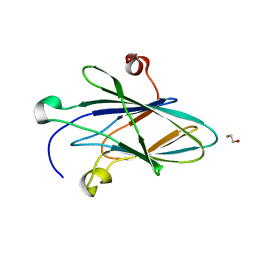 | | Crystal Structure of Myxoma Virus M64 | | Descriptor: | BETA-MERCAPTOETHANOL, M64R | | Authors: | Krumm, B.E, Meng, X, Li, Y, Xiang, Y, Deng, J. | | Deposit date: | 2015-07-31 | | Release date: | 2015-11-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for antagonizing a host restriction factor by C7 family of poxvirus host-range proteins.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4W60
 
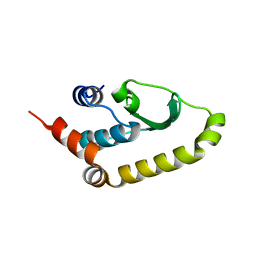 | | The structure of Vaccina virus H7 protein displays A Novel Phosphoinositide binding fold required for membrane biogenesis | | Descriptor: | Late protein H7 | | Authors: | Kolli, S, Meng, X, Wu, X, Shengjuler, D, Cameron, C.E, Xiang, Y, Deng, J. | | Deposit date: | 2014-08-19 | | Release date: | 2014-12-31 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-function analysis of vaccinia virus h7 protein reveals a novel phosphoinositide binding fold essential for poxvirus replication.
J.Virol., 89, 2015
|
|
4XFU
 
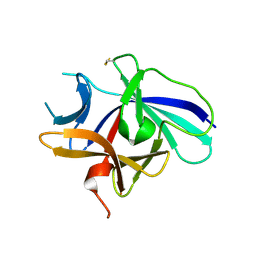 | | Structure of IL-18 SER Mutant V | | Descriptor: | Interleukin-18 | | Authors: | Krumm, B.E, Meng, X, Xiang, Y, Deng, J. | | Deposit date: | 2014-12-29 | | Release date: | 2015-06-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystallization of interleukin-18 for structure-based inhibitor design.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
4XFT
 
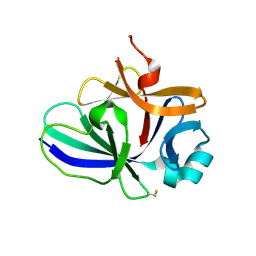 | | Structure of IL-18 SER Mutant III | | Descriptor: | DIMETHYL SULFOXIDE, Interleukin-18 | | Authors: | Krumm, B.E, Meng, X, Xiang, Y, Deng, J. | | Deposit date: | 2014-12-29 | | Release date: | 2015-06-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallization of interleukin-18 for structure-based inhibitor design.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
4XFS
 
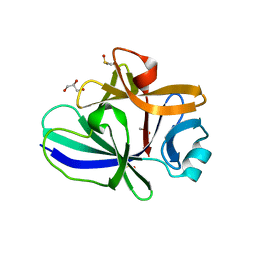 | | Structure of IL-18 SER Mutant I | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, Interleukin-18 | | Authors: | Krumm, B.E, Meng, X, Xiang, Y, Deng, J. | | Deposit date: | 2014-12-28 | | Release date: | 2015-06-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystallization of interleukin-18 for structure-based inhibitor design.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
6OMM
 
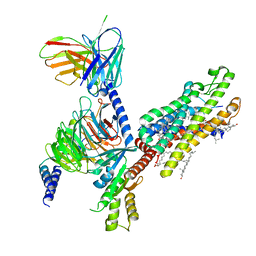 | | Cryo-EM structure of formyl peptide receptor 2/lipoxin A4 receptor in complex with Gi | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhuang, Y, Liu, H, de Waal, P.W, Zhou, X.E, Wang, L, Meng, X, Zhao, G, Kang, Y, Melcher, K, Xu, H.E, Zhang, C. | | Deposit date: | 2019-04-19 | | Release date: | 2020-02-26 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Structure of formylpeptide receptor 2-Gicomplex reveals insights into ligand recognition and signaling.
Nat Commun, 11, 2020
|
|
6PT0
 
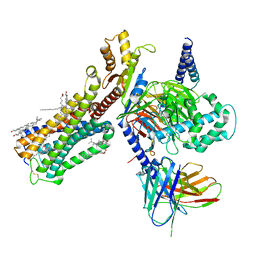 | | Cryo-EM structure of human cannabinoid receptor 2-Gi protein in complex with agonist WIN 55,212-2 | | Descriptor: | CHOLESTEROL, Cannabinoid receptor 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Xu, T.H, Xing, C, Zhuang, Y, Feng, Z, Zhou, X.E, Chen, M, Wang, L, Meng, X, Xue, Y, Wang, J, Liu, H, McGuire, T, Zhao, G, Melcher, K, Zhang, C, Xu, H.E, Xie, X.Q. | | Deposit date: | 2019-07-14 | | Release date: | 2020-02-12 | | Last modified: | 2020-03-04 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM Structure of the Human Cannabinoid Receptor CB2-GiSignaling Complex.
Cell, 180, 2020
|
|
3J4F
 
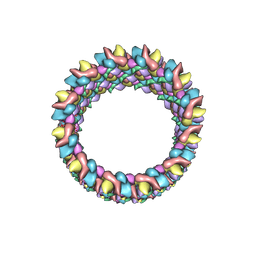 | | Structure of HIV-1 capsid protein by cryo-EM | | Descriptor: | capsid protein | | Authors: | Zhao, G, Perilla, J.R, Meng, X, Schulten, K, Zhang, P. | | Deposit date: | 2013-07-11 | | Release date: | 2013-07-24 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (8.6 Å) | | Cite: | Mature HIV-1 capsid structure by cryo-electron microscopy and all-atom molecular dynamics.
Nature, 497, 2013
|
|
3J34
 
 | | Structure of HIV-1 Capsid Protein by Cryo-EM | | Descriptor: | capsid protein | | Authors: | Zhao, G, Perilla, J.R, Yufenyuy, E, Meng, X, Chen, B, Ning, J, Ahn, J, Gronenborn, A.M, Schulten, K, Aiken, C, Zhang, P. | | Deposit date: | 2013-02-23 | | Release date: | 2013-05-29 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (8.6 Å) | | Cite: | Mature HIV-1 capsid structure by cryo-electron microscopy and all-atom molecular dynamics.
Nature, 497, 2013
|
|
6CMO
 
 | | Rhodopsin-Gi complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab Heavy chain, Fab light chain, ... | | Authors: | Kang, Y, Kuybeda, O, de Waal, P.W, Mukherjee, S, Van Eps, N, Dutka, P, Zhou, X.E, Bartesaghi, A, Erramilli, S, Morizumi, T, Gu, X, Yin, Y, Liu, P, Jiang, Y, Meng, X, Zhao, G, Melcher, K, Earnst, O.P, Kossiakoff, A.A, Subramaniam, S, Xu, H.E. | | Deposit date: | 2018-03-05 | | Release date: | 2018-06-20 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Cryo-EM structure of human rhodopsin bound to an inhibitory G protein.
Nature, 558, 2018
|
|
7VUX
 
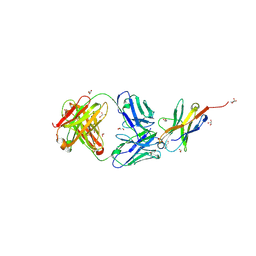 | | Complex structure of PD1 and 609A-Fab | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Huang, H, Zhu, Z, Zhao, J, Jiang, L, Yang, H, Deng, L, Meng, X, Ding, J, Yang, S, Zhao, L, Xu, W, Wang, X. | | Deposit date: | 2021-11-04 | | Release date: | 2021-11-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | A strategy for the efficient construction of anti-PD1-based bispecific antibodies with desired IgG-like properties.
Mabs, 14, 2022
|
|
6XA4
 
 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with UAW241 | | Descriptor: | 3C-like proteinase, GLYCEROL, inhibitor UAW241 | | Authors: | Sacco, M, Ma, C, Wang, J, Chen, Y. | | Deposit date: | 2020-06-03 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure and inhibition of the SARS-CoV-2 main protease reveal strategy for developing dual inhibitors against M pro and cathepsin L.
Sci Adv, 6, 2020
|
|
4W5X
 
 | |
4WUM
 
 | |
7RN0
 
 | |
7RN1
 
 | |
8CZ9
 
 | |
5IMX
 
 | |
4C13
 
 | | x-ray crystal structure of Staphylococcus aureus MurE with UDP-MurNAc- Ala-Glu-Lys | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Ruane, K.M, Roper, D.I, Fulop, V, Barreteau, H, Boniface, A, Dementin, S, Blanot, D, Mengin-Lecreulx, D, Gobec, S, Dessen, A, Dowson, C.G, Lloyd, A.J. | | Deposit date: | 2013-08-09 | | Release date: | 2013-10-02 | | Last modified: | 2021-03-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of a first-in-class CDK2 selective degrader for AML differentiation therapy.
Nat.Chem.Biol., 2021
|
|
5JLJ
 
 | | Crystal Structure of KPT8602 in complex with CRM1-Ran-RanBP1 | | Descriptor: | (2R)-3-{3-[3,5-bis(trifluoromethyl)phenyl]-1H-1,2,4-triazol-1-yl}-2-(pyrimidin-5-yl)propanamide, CHLORIDE ION, Exportin-1, ... | | Authors: | Fung, H.Y, Chook, Y.M. | | Deposit date: | 2016-04-27 | | Release date: | 2016-06-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Next-generation XPO1 inhibitor shows improved efficacy and in vivo tolerability in hematological malignancies.
Leukemia, 30, 2016
|
|
7KX5
 
 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with noncovalent inhibitor Jun8-76-3A | | Descriptor: | 3C-like proteinase, GLYCEROL, N-([1,1'-biphenyl]-4-yl)-N-[(1R)-2-oxo-2-{[(1S)-1-phenylethyl]amino}-1-(pyridin-3-yl)ethyl]furan-2-carboxamide | | Authors: | Sacco, M, Wang, J, Chen, Y. | | Deposit date: | 2020-12-03 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of Di- and Trihaloacetamides as Covalent SARS-CoV-2 Main Protease Inhibitors with High Target Specificity.
J.Am.Chem.Soc., 143, 2021
|
|
8HGH
 
 | | Structure of 2:2 PAPP-A.STC2 complex | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Pappalysin-1, Stanniocalcin-2, ZINC ION | | Authors: | Zhong, Q.H, Chu, H.L, Wang, G.P, Zhang, C, Wei, Y, Qiao, J, Hang, J. | | Deposit date: | 2022-11-14 | | Release date: | 2023-01-11 | | Method: | ELECTRON MICROSCOPY (4.16 Å) | | Cite: | Structural insights into the covalent regulation of PAPP-A activity by proMBP and STC2.
Cell Discov, 8, 2022
|
|
