3ERA
 
 | | RECOMBINANT ERABUTOXIN A (S8T MUTANT) | | Descriptor: | ERABUTOXIN A, THIOCYANATE ION | | Authors: | Gaucher, J.F, Menez, R, Arnoux, B, Menez, A, Ducruix, A. | | Deposit date: | 1997-06-25 | | Release date: | 1997-12-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High resolution x-ray analysis of two mutants of a curaremimetic snake toxin
Eur.J.Biochem., 267, 2000
|
|
1BGK
 
 | | SEA ANEMONE TOXIN (BGK) WITH HIGH AFFINITY FOR VOLTAGE DEPENDENT POTASSIUM CHANNEL, NMR, 15 STRUCTURES | | Descriptor: | BGK | | Authors: | Dauplais, M, Lecoq, A, Song, J, Cotton, J, Jamin, N, Gilquin, B, Roumestand, C, Vita, C, Harvey, A, Menez, A. | | Deposit date: | 1996-05-08 | | Release date: | 1997-01-27 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | On the convergent evolution of animal toxins. Conservation of a diad of functional residues in potassium channel-blocking toxins with unrelated structures.
J.Biol.Chem., 272, 1997
|
|
1BF0
 
 | | CALCICLUDINE (CAC) FROM GREEN MAMBA DENDROASPIS ANGUSTICEPS, NMR, 15 STRUCTURES | | Descriptor: | CALCICLUDINE | | Authors: | Gilquin, B, Lecoq, A, Desne, F, Guenneugues, M, Zinn-Justin, S, Menez, A. | | Deposit date: | 1998-05-26 | | Release date: | 1999-01-13 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | Conformational and functional variability supported by the BPTI fold: solution structure of the Ca2+ channel blocker calcicludine.
Proteins, 34, 1999
|
|
1QM7
 
 | | X-ray structure of a three-fingered chimeric protein, stability of a structural scaffold | | Descriptor: | R-CHII | | Authors: | Le Du, M.H, Ricciardi, A, Khayati, M, Menez, R, Boulain, J.C, Menez, A, Ducancel, F. | | Deposit date: | 1999-09-21 | | Release date: | 2000-03-15 | | Last modified: | 2017-07-05 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Stability of a Structural Scaffold Upon Activity Transfer : X-Ray Structure of a Three Fingers Chimeric Protein.
J.Mol.Biol., 296, 2000
|
|
1B45
 
 | | ALPHA-CNIA CONOTOXIN FROM CONUS CONSORS, NMR, 43 STRUCTURES | | Descriptor: | ALPHA-CNIA | | Authors: | Favreau, P, Krimm, I, Le Gall, F, Bobenrieth, M.J, Lamthanh, H, Bouet, F, Servent, D, Molgo, J, Menez, A, Letourneux, Y, Lancelin, J.M. | | Deposit date: | 1999-01-05 | | Release date: | 1999-07-09 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Biochemical characterization and nuclear magnetic resonance structure of novel alpha-conotoxins isolated from the venom of Conus consors.
Biochemistry, 38, 1999
|
|
1BAH
 
 | | A TWO DISULFIDE DERIVATIVE OF CHARYBDOTOXIN WITH DISULFIDE 13-33 REPLACED BY TWO ALPHA-AMINOBUTYRIC ACIDS, NMR, 30 STRUCTURES | | Descriptor: | CHARYBDOTOXIN | | Authors: | Song, J, Gilquin, B, Jamin, N, Guenneugues, M, Dauplais, M, Vita, C, Menez, A. | | Deposit date: | 1996-06-06 | | Release date: | 1997-01-11 | | Last modified: | 2020-01-15 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of a two-disulfide derivative of charybdotoxin: structural evidence for conservation of scorpion toxin alpha/beta motif and its hydrophobic side chain packing.
Biochemistry, 36, 1997
|
|
2ERA
 
 | | RECOMBINANT ERABUTOXIN A, S8G MUTANT | | Descriptor: | ERABUTOXIN A | | Authors: | Gaucher, J.F, Menez, R, Arnoux, B, Menez, A, Ducruix, A. | | Deposit date: | 1997-06-25 | | Release date: | 1997-12-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | High resolution x-ray analysis of two mutants of a curaremimetic snake toxin
Eur.J.Biochem., 267, 2000
|
|
2HEO
 
 | | General Structure-Based Approach to the Design of Protein Ligands: Application to the Design of Kv1.2 Potassium Channel Blockers. | | Descriptor: | 5'-D(*TP*CP*GP*CP*GP*CP*G)-3', Z-DNA binding protein 1 | | Authors: | Magis, C, Gasparini, S, Charbonnier, J.B, Stura, E, Le Du, M.H, Menez, A, Cuniasse, P. | | Deposit date: | 2006-06-21 | | Release date: | 2006-11-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-based secondary structure-independent approach to design protein ligands: Application to the design of Kv1.2 potassium channel blockers.
J.Am.Chem.Soc., 128, 2006
|
|
2VLW
 
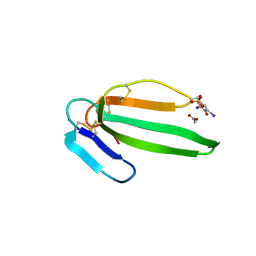 | | Crystal structure of the muscarinic toxin MT7 diiodoTYR51 derivative. | | Descriptor: | ACETATE ION, MUSCARINIC M1-TOXIN1, SULFATE ION | | Authors: | Menez, R, Granata, V, Mourier, G, Fruchart-Gaillard, C, Menez, A, Servant, D, Stura, E.A. | | Deposit date: | 2008-01-17 | | Release date: | 2008-10-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Different Interactions between Mt7 Toxin and the Human Muscarinic M1 Receptor in its Free and N-Methylscopolamine-Occupied States.
Mol.Pharmacol., 74, 2008
|
|
1FF4
 
 | |
1CMR
 
 | | NMR SOLUTION STRUCTURE OF A CHIMERIC PROTEIN, DESIGNED BY TRANSFERRING A FUNCTIONAL SNAKE BETA-HAIRPIN INTO A SCORPION ALPHA/BETA SCAFFOLD (PH 3.5, 20C), NMR, 18 STRUCTURES | | Descriptor: | CHARYBDOTOXIN, ALPHA CHIMERA | | Authors: | Zinn-Justin, S, Guenneugues, M, Drakopoulou, E, Gilquin, B, Vita, C, Menez, A. | | Deposit date: | 1996-03-15 | | Release date: | 1996-08-01 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | Transfer of a beta-hairpin from the functional site of snake curaremimetic toxins to the alpha/beta scaffold of scorpion toxins: three-dimensional solution structure of the chimeric protein.
Biochemistry, 35, 1996
|
|
1QUZ
 
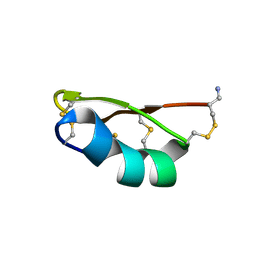 | | Solution structure of the potassium channel scorpion toxin HSTX1 | | Descriptor: | HSTX1 TOXIN | | Authors: | Savarin, P, Romi-Lebrun, R, Zinn-Justin, S, Lebrun, B, Nakajima, T, Gilquin, B, Menez, A. | | Deposit date: | 1999-07-05 | | Release date: | 2000-07-07 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Structural and functional consequences of the presence of a fourth disulfide bridge in the scorpion short toxins: solution structure of the potassium channel inhibitor HsTX1.
Protein Sci., 8, 1999
|
|
1CXN
 
 | | REFINED THREE-DIMENSIONAL SOLUTION STRUCTURE OF A SNAKE CARDIOTOXIN: ANALYSIS OF THE SIDE-CHAIN ORGANISATION SUGGESTS THE EXISTENCE OF A POSSIBLE PHOSPHOLIPID BINDING SITE | | Descriptor: | CARDIOTOXIN GAMMA | | Authors: | Gilquin, B, Roumestand, C, Zinn-Justin, S, Menez, A, Toma, F. | | Deposit date: | 1994-07-08 | | Release date: | 1994-12-20 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Refined three-dimensional solution structure of a snake cardiotoxin: analysis of the side-chain organization suggests the existence of a possible phospholipid binding site.
Biopolymers, 33, 1993
|
|
1NEA
 
 | | THREE-DIMENSIONAL SOLUTION STRUCTURE OF A CURAREMIMETIC TOXIN FROM NAJA NIGRICOLLIS VENOM: A PROTON NMR AND MOLECULAR MODELING STUDY | | Descriptor: | TOXIN ALPHA | | Authors: | Zinn-Justin, S, Roumestand, C, Gilquin, B, Bontems, F, Menez, A, Toma, F. | | Deposit date: | 1992-09-22 | | Release date: | 1993-10-31 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of a curaremimetic toxin from Naja nigricollis venom: a proton NMR and molecular modeling study.
Biochemistry, 31, 1992
|
|
1CXO
 
 | | REFINED THREE-DIMENSIONAL SOLUTION STRUCTURE OF A SNAKE CARDIOTOXIN: ANALYSIS OF THE SIDE-CHAIN ORGANISATION SUGGESTS THE EXISTENCE OF A POSSIBLE PHOSPHOLIPID BINDING SITE | | Descriptor: | CARDIOTOXIN GAMMA | | Authors: | Gilquin, B, Roumestand, C, Zinn-Justin, S, Menez, A, Toma, F. | | Deposit date: | 1994-11-07 | | Release date: | 1994-12-20 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Refined three-dimensional solution structure of a snake cardiotoxin: analysis of the side-chain organization suggests the existence of a possible phospholipid binding site.
Biopolymers, 33, 1993
|
|
1G2G
 
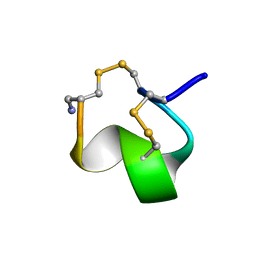 | | MINIMAL CONFORMATION OF THE ALPHA-CONOTOXIN IMI FOR THE ALPHA7 NEURONAL NICOTINIC ACETYLCHOLINE RECEPTOR RECOGNITION | | Descriptor: | ALPHA-CONOTOXIN IMI | | Authors: | Lamthanh, H, Jegou-Matheron, C, Servent, D, Menez, A, Lancelin, J.M. | | Deposit date: | 2000-10-19 | | Release date: | 2000-11-08 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Minimal conformation of the alpha-conotoxin ImI for the alpha7 neuronal nicotinic acetylcholine receptor recognition: correlated CD, NMR and binding studies.
FEBS Lett., 454, 1999
|
|
1D5Q
 
 | | SOLUTION STRUCTURE OF A MINI-PROTEIN REPRODUCING THE CORE OF THE CD4 SURFACE INTERACTING WITH THE HIV-1 ENVELOPE GLYCOPROTEIN | | Descriptor: | CHIMERIC MINI-PROTEIN | | Authors: | Vita, C, Drakopoulou, E, Vizzanova, J, Rochette, S, Martin, L, Menez, A, Roumestand, C, Yang, Y.S, Ylisastigui, L, Benjouad, A, Gluckman, J.C. | | Deposit date: | 1999-10-11 | | Release date: | 2000-10-11 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Rational engineering of a miniprotein that reproduces the core of the CD4 site interacting with HIV-1 envelope glycoprotein.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
2GLQ
 
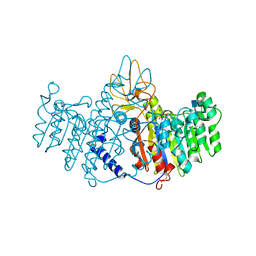 | | X-ray structure of human alkaline phosphatase in complex with strontium | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Alkaline phosphatase, placental type, ... | | Authors: | Llinas, P, Masella, M, Stigbrand, T, Menez, A, Stura, E.A, Le Du, M.H. | | Deposit date: | 2006-04-05 | | Release date: | 2006-04-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural studies of human alkaline phosphatase in complex with strontium: Implication for its secondary effect in bones.
Protein Sci., 15, 2006
|
|
1IQ9
 
 | |
1JNL
 
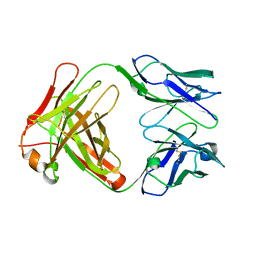 | | Crystal Structure of Fab-Estradiol Complexes | | Descriptor: | monoclonal anti-estradiol 17E12E5 immunoglobulin gamma-1 chain, monoclonal anti-estradiol 17E12E5 immunoglobulin kappa chain | | Authors: | Monnet, C, Bettsworth, F, Stura, E.A, Le Du, M.-H, Menez, R, Derrien, L, Zinn-Justin, S, Gilquin, B, Sibai, G, Battail-Poirot, N, Jolivet, M, Menez, A, Arnaud, M, Ducancel, F, Charbonnier, J.B. | | Deposit date: | 2001-07-24 | | Release date: | 2002-02-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Highly specific anti-estradiol antibodies: structural characterisation and binding diversity.
J.Mol.Biol., 315, 2002
|
|
1JN6
 
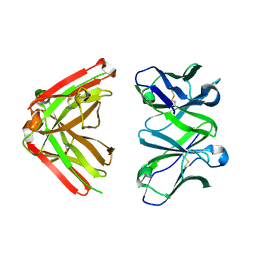 | | Crystal Structure of Fab-Estradiol Complexes | | Descriptor: | monoclonal anti-estradiol 10G6D6 Fab heavy chain, monoclonal anti-estradiol 10G6D6 Fab light chain | | Authors: | Monnet, C, Bettsworth, F, Stura, E.A, Le Du, M.-H, Menez, R, Derrien, L, Zinn-Justin, S, Gilquin, B, Sibai, G, Battail-Poirot, N, Jolivet, M, Menez, A, Arnaud, M, Ducancel, F, Charbonnier, J.B. | | Deposit date: | 2001-07-23 | | Release date: | 2002-02-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Highly specific anti-estradiol antibodies: structural characterisation and binding diversity.
J.Mol.Biol., 315, 2002
|
|
1JNN
 
 | | Crystal Structure of Fab-Estradiol Complexes | | Descriptor: | ESTRADIOL, MONOCLONAL ANTI-ESTRADIOL 17E12E5 IMMUNOGLOBULIN GAMMA-1 CHAIN, MONOCLONAL ANTI-ESTRADIOL 17E12E5 IMMUNOGLOBULIN KAPPA CHAIN | | Authors: | Monnet, C, Bettsworth, F, Stura, E.A, Le Du, M.-H, Menez, R, Derrien, L, Zinn-Justin, S, Gilquin, B, Sibai, G, Battail-Poirot, N, Jolivet, M, Menez, A, Arnaud, M, Ducancel, F, Charbonnier, J.B. | | Deposit date: | 2001-07-24 | | Release date: | 2002-02-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Highly specific anti-estradiol antibodies: structural characterisation and binding diversity.
J.Mol.Biol., 315, 2002
|
|
1JNH
 
 | | Crystal Structure of Fab-Estradiol Complexes | | Descriptor: | ESTRADIOL-6 CARBOXYL-METHYL-OXIME, monoclonal anti-estradiol 10G6D6 Fab heavy chain, monoclonal anti-estradiol 10G6D6 Fab light chain | | Authors: | Monnet, C, Bettsworth, F, Stura, E.A, Le Du, M.-H, Menez, R, Derrien, L, Zinn-Justin, S, Gilquin, B, Sibai, G, Battail-Poirot, N, Jolivet, M, Menez, A, Arnaud, M, Ducancel, F, Charbonnier, J.B. | | Deposit date: | 2001-07-24 | | Release date: | 2002-02-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Highly specific anti-estradiol antibodies: structural characterisation and binding diversity.
J.Mol.Biol., 315, 2002
|
|
1SXM
 
 | | SCORPION TOXIN (NOXIUSTOXIN) WITH HIGH AFFINITY FOR VOLTAGE DEPENDENT POTASSIUM CHANNEL AND LOW AFFINITY FOR CALCIUM DEPENDENT POTASSIUM CHANNEL (NMR AT 20 DEGREES, PH3.5, 39 STRUCTURES) | | Descriptor: | NOXIUSTOXIN | | Authors: | Dauplais, M, Gilquin, B, Possani, L.D, Gurrola-Briones, G, Roumestand, C, Menez, A. | | Deposit date: | 1995-09-07 | | Release date: | 1996-01-29 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Determination of the three-dimensional solution structure of noxiustoxin: analysis of structural differences with related short-chain scorpion toxins.
Biochemistry, 34, 1995
|
|
1EW2
 
 | | CRYSTAL STRUCTURE OF A HUMAN PHOSPHATASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, PHOSPHATASE, ... | | Authors: | Le Du, M.H, Stigbrand, T, Taussig, M.J, Menez, A, Stura, E.A. | | Deposit date: | 2000-04-21 | | Release date: | 2001-04-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structure of alkaline phosphatase from human placenta at 1.8 A resolution. Implication for a substrate specificity.
J.Biol.Chem., 276, 2001
|
|
