1EJB
 
 | | LUMAZINE SYNTHASE FROM SACCHAROMYCES CEREVISIAE | | Descriptor: | 5-(6-D-RIBITYLAMINO-2,4-DIHYDROXYPYRIMIDIN-5-YL)-1-PENTYL-PHOSPHONIC ACID, LUMAZINE SYNTHASE | | Authors: | Meining, W, Mortl, S, Fischer, M, Cushman, M, Bacher, A, Ladenstein, R. | | Deposit date: | 2000-03-02 | | Release date: | 2001-03-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The atomic structure of pentameric lumazine synthase from Saccharomyces cerevisiae at 1.85 A resolution reveals the binding mode of a phosphonate intermediate analogue.
J.Mol.Biol., 299, 2000
|
|
3QKG
 
 | |
1PKV
 
 | | The N-terminal domain of riboflavin synthase in complex with riboflavin | | Descriptor: | RIBOFLAVIN, Riboflavin synthase alpha chain | | Authors: | Meining, W, Eberhardt, S, Bacher, A, Ladenstein, R. | | Deposit date: | 2003-06-06 | | Release date: | 2004-06-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The structure of the N-terminal domain of riboflavin synthase in complex with riboflavin at 2.6A resolution.
J.Mol.Biol., 331, 2003
|
|
8B50
 
 | |
2C4E
 
 | | Crystal Structure of Methanocaldococcus jannaschii Nucleoside Kinase - An Archaeal Member of the Ribokinase Family | | Descriptor: | MAGNESIUM ION, SUGAR KINASE MJ0406 | | Authors: | Arnfors, L, Hansen, T, Meining, W, Schoenheit, P, Ladenstein, R. | | Deposit date: | 2005-10-18 | | Release date: | 2006-08-30 | | Last modified: | 2018-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of Methanocaldococcus Jannaschii Nucleoside Kinase: An Archaeal Member of the Ribokinase Family.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2C49
 
 | | Crystal Structure of Methanocaldococcus jannaschii Nucleoside Kinase - An Archaeal Member of the Ribokinase Family | | Descriptor: | ADENOSINE, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Arnfors, L, Hansen, T, Meining, W, Schoenheit, P, Ladenstein, R. | | Deposit date: | 2005-10-17 | | Release date: | 2006-08-30 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structure of Methanocaldococcus Jannaschii Nucleoside Kinase: An Archaeal Member of the Ribokinase Family.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
1W29
 
 | | Lumazine Synthase from Mycobacterium tuberculosis bound to 3-(1,3,7- trihydro-9-D-ribityl-2,6,8-purinetrione-7-yl)butane 1-phosphate | | Descriptor: | (4S,5S)-1,2-DITHIANE-4,5-DIOL, 4-{2,6,8-TRIOXO-9-[(2R,3S,4R)-2,3,4,5-TETRAHYDROXYPENTYL]-1,2,3,6,8,9-HEXAHYDRO-7H-PURIN-7-YL}BUTYL DIHYDROGEN PHOSPHATE, 4-{2,6,8-TRIOXO-9-[(2S,3R,4R)-2,3,4,5-TETRAHYDROXYPENTYL]-1,2,3,6,8,9-HEXAHYDRO-7H-PURIN-7-YL}BUTYL DIHYDROGEN PHOSPHATE, ... | | Authors: | Morgunova, E, Meining, W, Illarionov, B, Haase, I, Fischer, M, Cushman, M, Bacher, A, Ladenstein, R. | | Deposit date: | 2004-07-01 | | Release date: | 2005-03-03 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Lumazine Synthase from Mycobacterium Tuberculosis as a Target for Rational Drug Design: Binding Mode of a New Class of Purinetrione Inhibitors(,)
Biochemistry, 44, 2005
|
|
1W19
 
 | | Lumazine Synthase from Mycobacterium tuberculosis bound to 3-(1,3,7- trihydro-9-D-ribityl-2,6,8-purinetrione-7-yl)propane 1-phosphate | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, (4S,5S)-1,2-DITHIANE-4,5-DIOL, ... | | Authors: | Morgunova, E, Meining, W, Illarionov, B, Haase, I, Fischer, M, Cushman, M, Bacher, A, Ladenstein, R. | | Deposit date: | 2004-06-03 | | Release date: | 2005-03-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Lumazine Synthase from Mycobacterium Tuberculosis as a Target for Rational Drug Design: Binding Mode of a New Class of Purinetrione Inhibitors(,)
Biochemistry, 44, 2005
|
|
1GTP
 
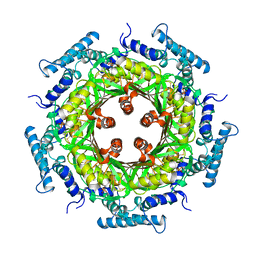 | | GTP CYCLOHYDROLASE I | | Descriptor: | GTP CYCLOHYDROLASE I, SULFATE ION | | Authors: | Nar, H, Huber, R, Meining, W, Bacher, A. | | Deposit date: | 1995-09-16 | | Release date: | 1996-11-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Atomic structure of GTP cyclohydrolase I.
Structure, 3, 1995
|
|
1HQK
 
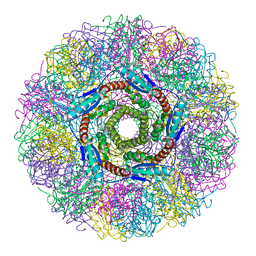 | | CRYSTAL STRUCTURE ANALYSIS OF LUMAZINE SYNTHASE FROM AQUIFEX AEOLICUS | | Descriptor: | 6,7-DIMETHYL-8-RIBITYLLUMAZINE SYNTHASE | | Authors: | Zhang, X, Meining, W, Fischer, M, Bacher, A, Ladenstein, R. | | Deposit date: | 2000-12-18 | | Release date: | 2001-12-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray structure analysis and crystallographic refinement of lumazine synthase from the hyperthermophile Aquifex aeolicus at 1.6 A resolution: determinants of thermostability revealed from structural comparisons.
J.Mol.Biol., 306, 2001
|
|
4I5E
 
 | | Crystal structure of Ralstonia sp. alcohol dehydrogenase in complex with NADP+ | | Descriptor: | Alclohol dehydrogenase/short-chain dehydrogenase, GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Jarasch, A, Lerchner, A, Meining, W, Schiefner, A, Skerra, A. | | Deposit date: | 2012-11-28 | | Release date: | 2013-06-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystallographic analysis and structure-guided engineering of NADPH-dependent Ralstonia sp. Alcohol dehydrogenase toward NADH cosubstrate specificity.
Biotechnol.Bioeng., 110, 2013
|
|
4I5D
 
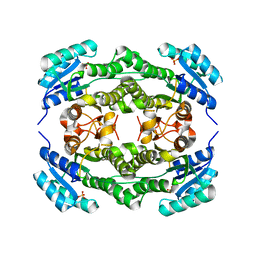 | | Crystal structure of Ralstonia sp. alcohol dehydrogenase in its apo form | | Descriptor: | Alclohol dehydrogenase/short-chain dehydrogenase, SULFATE ION | | Authors: | Jarasch, A, Lerchner, A, Meining, W, Schiefner, A, Skerra, A. | | Deposit date: | 2012-11-28 | | Release date: | 2013-06-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystallographic analysis and structure-guided engineering of NADPH-dependent Ralstonia sp. Alcohol dehydrogenase toward NADH cosubstrate specificity.
Biotechnol.Bioeng., 110, 2013
|
|
4I5F
 
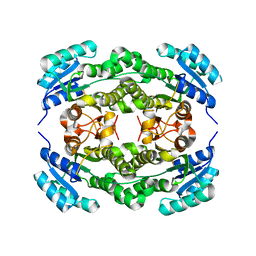 | | Crystal structure of Ralstonia sp. alcohol dehydrogenase mutant N15G, G37D, R38V, R39S | | Descriptor: | Alclohol dehydrogenase/short-chain dehydrogenase | | Authors: | Jarasch, A, Lerchner, A, Meining, W, Schiefner, A, Skerra, A. | | Deposit date: | 2012-11-28 | | Release date: | 2013-06-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystallographic analysis and structure-guided engineering of NADPH-dependent Ralstonia sp. Alcohol dehydrogenase toward NADH cosubstrate specificity.
Biotechnol.Bioeng., 110, 2013
|
|
4I5G
 
 | | Crystal structure of Ralstonia sp. alcohol dehydrogenase mutant N15G, G37D, R38V, R39S, A86N, S88A | | Descriptor: | Alclohol dehydrogenase/short-chain dehydrogenase | | Authors: | Jarasch, A, Lerchner, A, Meining, W, Schiefner, A, Skerra, A. | | Deposit date: | 2012-11-28 | | Release date: | 2013-06-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallographic analysis and structure-guided engineering of NADPH-dependent Ralstonia sp. Alcohol dehydrogenase toward NADH cosubstrate specificity.
Biotechnol.Bioeng., 110, 2013
|
|
1NQV
 
 | | Crystal Structure of Lumazine Synthase from Aquifex aeolicus in Complex with Inhibitor: 5-nitroso-6-ribityl-amino-2,4(1H,3H)pyrimidinedione | | Descriptor: | 5-NITROSO-6-RIBITYL-AMINO-2,4(1H,3H)-PYRIMIDINEDIONE, 6,7-dimethyl-8-ribityllumazine synthase, PHOSPHATE ION | | Authors: | Zhang, X, Meining, W, Cushman, M, Haase, I, Fischer, M, Bacher, A, Ladenstein, R. | | Deposit date: | 2003-01-23 | | Release date: | 2004-01-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | A structure-based model of the reaction catalyzed by lumazine synthase from Aquifex aeolicus.
J.Mol.Biol., 328, 2003
|
|
1NQX
 
 | | Crystal Structure of Lumazine Synthase from Aquifex aeolicus in Complex with Inhibitor: 3-(7-hydroxy-8-ribityllumazine-6-yl)propionic acid | | Descriptor: | 3-(7-HYDROXY-8-RIBITYLLUMAZINE-6-YL) PROPIONIC ACID, 6,7-dimethyl-8-ribityllumazine synthase, PHOSPHATE ION | | Authors: | Zhang, X, Meining, W, Cushman, M, Haase, I, Fischer, M, Bacher, A, Ladenstein, R. | | Deposit date: | 2003-01-23 | | Release date: | 2004-01-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | A structure-based model of the reaction catalyzed by lumazine synthase from Aquifex aeolicus.
J.Mol.Biol., 328, 2003
|
|
1NQW
 
 | | Crystal Structure of Lumazine Synthase from Aquifex aeolicus in Complex with Inhibitor: 5-(6-D-ribitylamino-2,4(1H,3H)pyrimidinedione-5-yl)-1-pentyl-phosphonic acid | | Descriptor: | 5-(6-D-RIBITYLAMINO-2,4(1H,3H)PYRIMIDINEDIONE-5-YL) PENTYL-1-PHOSPHONIC ACID, 6,7-dimethyl-8-ribityllumazine synthase | | Authors: | Zhang, X, Meining, W, Cushman, M, Haase, I, Fischer, M, Bacher, A, Ladenstein, R. | | Deposit date: | 2003-01-23 | | Release date: | 2004-01-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A structure-based model of the reaction catalyzed by lumazine synthase from Aquifex aeolicus.
J.Mol.Biol., 328, 2003
|
|
1NQU
 
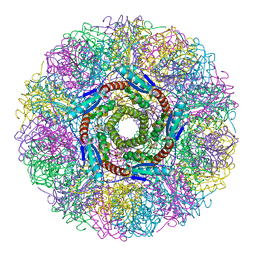 | | Crystal Structure of Lumazine Synthase from Aquifex aeolicus in Complex with Inhibitor: 6,7-dioxo-5H-8-ribitylaminolumazine | | Descriptor: | 6,7-DIOXO-5H-8-RIBITYLAMINOLUMAZINE, 6,7-dimethyl-8-ribityllumazine synthase, PHOSPHATE ION | | Authors: | Zhang, X, Meining, W, Cushman, M, Haase, I, Fischer, M, Bacher, A, Ladenstein, R. | | Deposit date: | 2003-01-23 | | Release date: | 2004-01-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A structure-based model of the reaction catalyzed by lumazine synthase from Aquifex aeolicus.
J.Mol.Biol., 328, 2003
|
|
1A9C
 
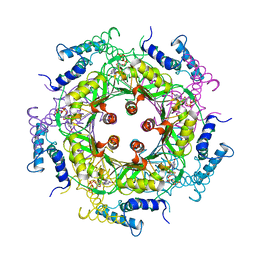 | | GTP CYCLOHYDROLASE I (C110S MUTANT) IN COMPLEX WITH GTP | | Descriptor: | GTP CYCLOHYDROLASE I, GUANOSINE-5'-TRIPHOSPHATE | | Authors: | Auerbach, G, Nar, H, Bracher, A, Bacher, A, Huber, R. | | Deposit date: | 1998-04-04 | | Release date: | 1999-05-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | GTP Cyclohydrolase I in Complex with GTP at 2.1 A Resolution
To be Published
|
|
1A8R
 
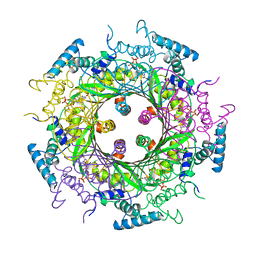 | | GTP CYCLOHYDROLASE I (H112S MUTANT) IN COMPLEX WITH GTP | | Descriptor: | GTP CYCLOHYDROLASE I, GUANOSINE-5'-TRIPHOSPHATE | | Authors: | Auerbach, G, Nar, H, Bracher, A, Bacher, A, Huber, R. | | Deposit date: | 1998-03-27 | | Release date: | 1999-05-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Biosynthesis of pteridines. Reaction mechanism of GTP cyclohydrolase I.
J.Mol.Biol., 326, 2003
|
|
1OAA
 
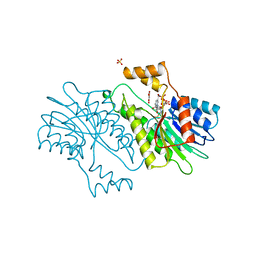 | | MOUSE SEPIAPTERIN REDUCTASE COMPLEXED WITH NADP AND OXALOACETATE | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, OXALOACETATE ION, SEPIAPTERIN REDUCTASE, ... | | Authors: | Auerbach, G, Herrmann, A, Bacher, A, Huber, R. | | Deposit date: | 1997-08-25 | | Release date: | 1999-02-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | The 1.25 A crystal structure of sepiapterin reductase reveals its binding mode to pterins and brain neurotransmitters.
EMBO J., 16, 1997
|
|
