4NZL
 
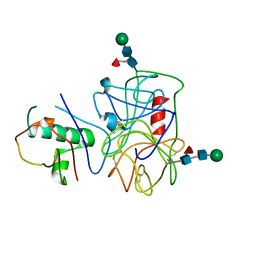 | | Extracellular proteins of Staphylococcus aureus inhibit the neutrophil serine proteases | | Descriptor: | Neutrophil elastase, Uncharacterized protein, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Stapels, D.A.C, von Koeckritz-Blickwede, M, Ramyar, K.X, Bischoff, M, Milder, F, Ruyken, M, Scheepmaker, L, McWhorter, W.J, Herrmann, M, van Kessel, K.P.M, Geisbrecht, B.V, Rooijakkers, S.H.M. | | Deposit date: | 2013-12-12 | | Release date: | 2014-08-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Staphylococcus aureus secretes a unique class of neutrophil serine protease inhibitors.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3V4X
 
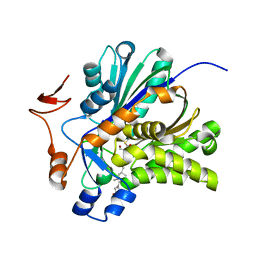 | | The Biochemical and Structural Basis for Inhibition of Enterococcus faecalis HMG-CoA Synthase, mvaS, by Hymeglusin | | Descriptor: | (7R,12R,13R)-13-formyl-12,14-dihydroxy-3,5,7-trimethyltetradeca-2,4-dienoic acid, HMG-CoA synthase | | Authors: | Skaff, D.A, Ramyar, K.X, McWhorter, W.J, Geisbrecht, B.V, Miziorko, H.M. | | Deposit date: | 2011-12-15 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | Biochemical and structural basis for inhibition of Enterococcus faecalis hydroxymethylglutaryl-CoA synthase, mvaS, by hymeglusin.
Biochemistry, 51, 2012
|
|
3V4N
 
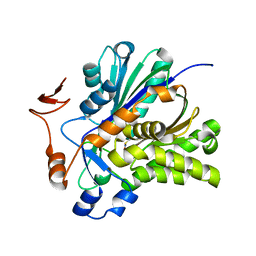 | | The Biochemical and Structural Basis for Inhibition of Enterococcus faecalis HMG-CoA Synthatse, mvaS, by Hymeglusin | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, HMG-CoA synthase | | Authors: | Skaff, D.A, Ramyar, K.X, McWhorter, W.J, Geisbrecht, B.V, Miziorko, H.M. | | Deposit date: | 2011-12-15 | | Release date: | 2012-04-25 | | Last modified: | 2014-09-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Biochemical and structural basis for inhibition of Enterococcus faecalis hydroxymethylglutaryl-CoA synthase, mvaS, by hymeglusin.
Biochemistry, 51, 2012
|
|
3GT2
 
 | | Crystal Structure of the P60 Domain from M. avium paratuberculosis Antigen MAP1272c | | Descriptor: | 1,2-ETHANEDIOL, Putative uncharacterized protein, SULFATE ION | | Authors: | Ramyar, K.X, Lingle, C.K, McWhorter, W.J, Bouyain, S, Bannantine, J.P, Geisbrecht, B.V. | | Deposit date: | 2009-03-27 | | Release date: | 2010-04-07 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structures of Two P60-Family Antigens from Mycobacterium Avium Paratuberculosis
To be Published
|
|
3I86
 
 | | Crystal structure of the P60 Domain from M. avium subspecies paratuberculosis antigen MAP1204 | | Descriptor: | ISOPROPYL ALCOHOL, Putative uncharacterized protein | | Authors: | Ramyar, K.X, Lingle, C.K, McWhorter, W.J, Bouyain, S, Bannantine, J.P, Geisbrecht, B.V. | | Deposit date: | 2009-07-09 | | Release date: | 2010-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Atypical structural features of two P60 family members from Mycobacterium avium subspecies paratuberculosis
To be Published
|
|
4DPU
 
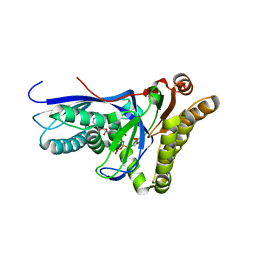 | |
4DPX
 
 | |
4DPT
 
 | | Crystal structure of Staphylococcus epidermidis mevalonate diphosphate decarboxylase complexed with inhibitor 6-FMVAPP and ATPgS | | Descriptor: | (3R)-3-(fluoromethyl)-3-hydroxy-5-{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}pentanoic acid, GLYCEROL, Mevalonate diphosphate decarboxylase, ... | | Authors: | Barta, M.L, McWhorter, W.J, Geisbrecht, B.V. | | Deposit date: | 2012-02-14 | | Release date: | 2012-07-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.191 Å) | | Cite: | Structural basis for nucleotide binding and reaction catalysis in mevalonate diphosphate decarboxylase.
Biochemistry, 51, 2012
|
|
4DU8
 
 | |
4DPW
 
 | | Crystal structure of Staphylococcus epidermidis D283A mevalonate diphosphate decarboxylase complexed with mevalonate diphosphate and ATPgS | | Descriptor: | (3R)-3-HYDROXY-5-{[(R)-HYDROXY(PHOSPHONOOXY)PHOSPHORYL]OXY}-3-METHYLPENTANOIC ACID, FORMIC ACID, GLYCEROL, ... | | Authors: | Barta, M.L, McWhorter, W.J, Geisbrecht, B.V. | | Deposit date: | 2012-02-14 | | Release date: | 2012-07-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.605 Å) | | Cite: | Structural basis for nucleotide binding and reaction catalysis in mevalonate diphosphate decarboxylase.
Biochemistry, 51, 2012
|
|
4DPY
 
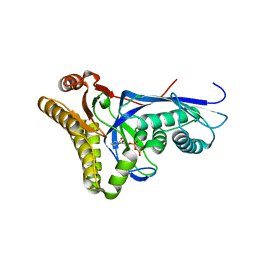 | |
4DU7
 
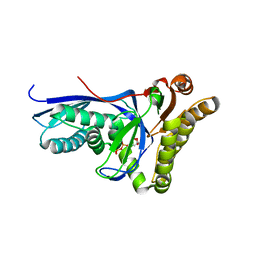 | |
3QT5
 
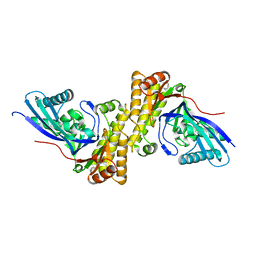 | | Crystal structure of Staphylococcus epidermidis mevalonate diphosphate decarboxylase | | Descriptor: | Mevalonate diphosphate decarboxylase | | Authors: | Barta, M.L, Skaff, A.D, McWhorter, W.J, Miziorko, H.M, Geisbrecht, B.V. | | Deposit date: | 2011-02-22 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.848 Å) | | Cite: | Crystal structures of Staphylococcus epidermidis mevalonate diphosphate decarboxylase bound to inhibitory analogs reveal new insight into substrate binding and catalysis.
J.Biol.Chem., 286, 2011
|
|
3QT6
 
 | | Crystal structure of Staphylococcus epidermidis mevalonate diphosphate decarboxylase complexed with inhibitor DPGP | | Descriptor: | 1-({[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}acetyl)-L-proline, Mevalonate diphosphate decarboxylase | | Authors: | Barta, M.L, Skaff, A.D, McWhorter, W.J, Miziorko, H.M, Geisbrecht, B.V. | | Deposit date: | 2011-02-22 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.047 Å) | | Cite: | Crystal structures of Staphylococcus epidermidis mevalonate diphosphate decarboxylase bound to inhibitory analogs reveal new insight into substrate binding and catalysis.
J.Biol.Chem., 286, 2011
|
|
3QT8
 
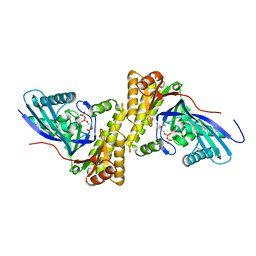 | | Crystal structure of mutant S192A Staphylococcus epidermidis mevalonate diphosphate decarboxylase complexed with inhibitor 6-FMVAPP | | Descriptor: | (3R)-3-(fluoromethyl)-3-hydroxy-5-{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}pentanoic acid, GLYCEROL, Mevalonate diphosphate decarboxylase | | Authors: | Barta, M.L, Skaff, A.D, McWhorter, W.J, Miziorko, H.M, Geisbrecht, B.V. | | Deposit date: | 2011-02-22 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of Staphylococcus epidermidis mevalonate diphosphate decarboxylase bound to inhibitory analogs reveal new insight into substrate binding and catalysis.
J.Biol.Chem., 286, 2011
|
|
3QT7
 
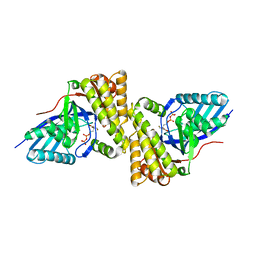 | | Crystal structure of Staphylococcus epidermidis mevalonate diphosphate decarboxylase complexed with inhibitor 6-FMVAPP | | Descriptor: | (3R)-3-(fluoromethyl)-3-hydroxy-5-{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}pentanoic acid, Mevalonate diphosphate decarboxylase | | Authors: | Barta, M.L, Skaff, A.D, McWhorter, W.J, Miziorko, H.M, Geisbrecht, B.V. | | Deposit date: | 2011-02-22 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | Crystal structures of Staphylococcus epidermidis mevalonate diphosphate decarboxylase bound to inhibitory analogs reveal new insight into substrate binding and catalysis.
J.Biol.Chem., 286, 2011
|
|
3L3O
 
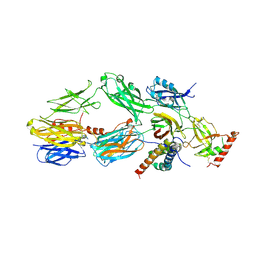 | |
3L5N
 
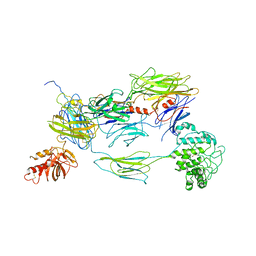 | |
3NMS
 
 | |
3OHX
 
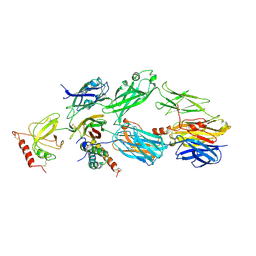 | |
6D69
 
 | |
