3SEA
 
 | | Structure of Rheb-Y35A mutant in GDP- and GMPPNP-bound forms | | Descriptor: | ACETATE ION, GTP-binding protein Rheb, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Mazhab-Jafari, M.T, Marshall, C.B, Ishiyama, N, Vuk, S, Ikura, M. | | Deposit date: | 2011-06-10 | | Release date: | 2012-06-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An Autoinhibited Noncanonical Mechanism of GTP Hydrolysis by Rheb Maintains mTORC1 Homeostasis.
Structure, 20, 2012
|
|
4O25
 
 | | Structure of Wild Type Mus musculus Rheb bound to GTP | | Descriptor: | GTP-binding protein Rheb, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION | | Authors: | Mazhab-Jafari, M.T, Marshall, C.B, Ho, J, Ishiyama, N, Stambolic, V, Ikura, M. | | Deposit date: | 2013-12-16 | | Release date: | 2014-03-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-guided mutation of the conserved G3-box glycine in Rheb generates a constitutively activated regulator of mammalian target of rapamycin (mTOR).
J.Biol.Chem., 289, 2014
|
|
4O2L
 
 | | Structure of Mus musculus Rheb G63A mutant bound to GTP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, GTP-binding protein Rheb, ... | | Authors: | Mazhab-Jafari, M.T, Marshall, C.B, Ho, J, Ishiyama, N, Stambolic, V, Ikura, M. | | Deposit date: | 2013-12-17 | | Release date: | 2014-03-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-guided mutation of the conserved G3-box glycine in Rheb generates a constitutively activated regulator of mammalian target of rapamycin (mTOR).
J.Biol.Chem., 289, 2014
|
|
4O2R
 
 | | Structure of Mus musculus Rheb G63V mutant bound to GDP | | Descriptor: | GTP-binding protein Rheb, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Mazhab-Jafari, M.T, Marshall, C.B, Ho, J, Ishiyama, N, Stambolic, V, Ikura, M. | | Deposit date: | 2013-12-17 | | Release date: | 2014-03-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-guided mutation of the conserved G3-box glycine in Rheb generates a constitutively activated regulator of mammalian target of rapamycin (mTOR).
J.Biol.Chem., 289, 2014
|
|
5TJ5
 
 | | Atomic model for the membrane-embedded motor of a eukaryotic V-ATPase | | Descriptor: | V-type proton ATPase subunit a, V-type proton ATPase subunit c, V-type proton ATPase subunit c', ... | | Authors: | Mazhab-Jafari, M.T, Rohou, A, Schmidt, C, Bueler, S.A, Benlekbir, S, Robinson, C.V, Rubinstein, J.L. | | Deposit date: | 2016-10-03 | | Release date: | 2016-10-26 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Atomic model for the membrane-embedded VO motor of a eukaryotic V-ATPase.
Nature, 539, 2016
|
|
7TUI
 
 | |
8GKC
 
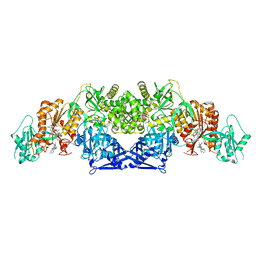 | |
7KDV
 
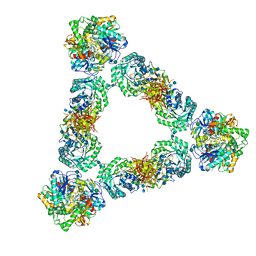 | | Murine core lysosomal multienzyme complex (LMC) composed of acid beta-galactosidase (GLB1) and protective protein cathepsin A (PPCA, CTSA) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-galactosidase, ... | | Authors: | Gorelik, A, Illes, K, Hasan, S.M.N, Nagar, B, Mazhab-Jafari, M.T. | | Deposit date: | 2020-10-09 | | Release date: | 2021-03-17 | | Last modified: | 2021-05-26 | | Method: | ELECTRON MICROSCOPY (4.59 Å) | | Cite: | Structure of the murine lysosomal multienzyme complex core.
Sci Adv, 7, 2021
|
|
8EYK
 
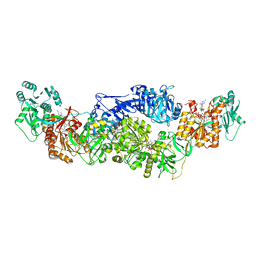 | |
8EYI
 
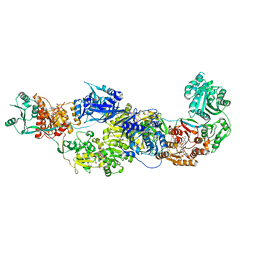 | |
8T7T
 
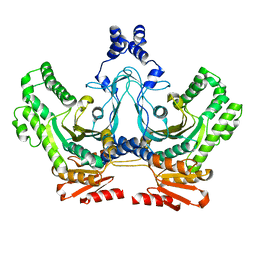 | |
6U5T
 
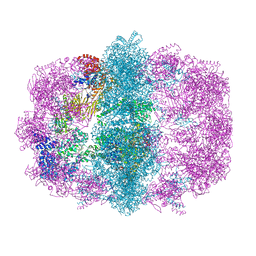 | |
6U5U
 
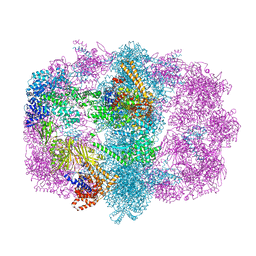 | |
6U5V
 
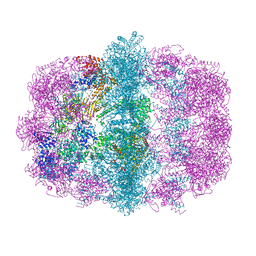 | |
6U5W
 
 | |
6WC7
 
 | |
2MSD
 
 | | NMR data-driven model of GTPase KRas-GNP tethered to a lipid-bilayer nanodisc | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Apolipoprotein A-I, GTPase KRas, ... | | Authors: | Mazhab-Jafari, M, Stathopoulos, P, Marshall, C, Ikura, M. | | Deposit date: | 2014-07-29 | | Release date: | 2015-06-03 | | Last modified: | 2019-12-11 | | Method: | SOLUTION NMR | | Cite: | Oncogenic and RASopathy-associated K-RAS mutations relieve membrane-dependent occlusion of the effector-binding site.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2MSE
 
 | | NMR data-driven model of GTPase KRas-GNP:ARafRBD complex tethered to a lipid-bilayer nanodisc | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Apolipoprotein A-I, GTPase KRas, ... | | Authors: | Mazhab-Jafari, M, Stathopoulos, P, Marshall, C, Ikura, M. | | Deposit date: | 2014-07-29 | | Release date: | 2015-06-03 | | Last modified: | 2019-12-11 | | Method: | SOLUTION NMR | | Cite: | Oncogenic and RASopathy-associated K-RAS mutations relieve membrane-dependent occlusion of the effector-binding site.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2MSC
 
 | | NMR data-driven model of GTPase KRas-GDP tethered to a lipid-bilayer nanodisc | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Apolipoprotein A-I, GTPase KRas, ... | | Authors: | Mazhab-Jafari, M, Stathopoulos, P, Marshall, C, Ikura, M. | | Deposit date: | 2014-07-29 | | Release date: | 2015-06-03 | | Last modified: | 2019-12-11 | | Method: | SOLUTION NMR | | Cite: | Oncogenic and RASopathy-associated K-RAS mutations relieve membrane-dependent occlusion of the effector-binding site.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
8DU5
 
 | | Murine sialidase-1 (NEU1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Sialidase-1, ... | | Authors: | Gorelik, A, Illes, K, Nagar, B. | | Deposit date: | 2022-07-26 | | Release date: | 2023-05-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of the immunoregulatory sialidase NEU1.
Sci Adv, 9, 2023
|
|
8U7Z
 
 | | KCTD5/Cullin3/Gbeta1gamma2 Complex: Local Refinment of KCTD5(CTD)/Gbeta1gamma2 | | Descriptor: | BTB/POZ domain-containing protein KCTD5, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 | | Authors: | Kuntz, D.A, Nguyen, D.M, Narayanan, N, Prive, G.G. | | Deposit date: | 2023-09-15 | | Release date: | 2023-10-11 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structure and dynamics of a pentameric KCTD5/Cullin3/G beta gamma E3 ubiquitin ligase complex
Biorxiv, 2023
|
|
8U83
 
 | | KCTD5/Cullin3/Gbeta1gamma2 Complex: State C From Composite RELION Multi-body Refinement Map | | Descriptor: | BTB/POZ domain-containing protein KCTD5, Cullin-3, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Kuntz, D.A, Nguyen, D.M, Narayanan, N, Prive, G.G. | | Deposit date: | 2023-09-15 | | Release date: | 2023-10-11 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3.975 Å) | | Cite: | Structure and dynamics of a pentameric KCTD5/Cullin3/G beta gamma E3 ubiquitin ligase complex
Biorxiv, 2023
|
|
8U80
 
 | |
8U81
 
 | | KCTD5/Cullin3/Gbeta1gamma2 Complex: State A From Composite RELION Multi-body Refinement Map | | Descriptor: | BTB/POZ domain-containing protein KCTD5, Cullin-3, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Kuntz, D.A, Nguyen, D.M, Narayanan, N, Prive, G.G. | | Deposit date: | 2023-09-15 | | Release date: | 2023-10-11 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3.82 Å) | | Cite: | Structure and dynamics of a pentameric KCTD5/Cullin3/G beta gamma E3 ubiquitin ligase complex
Biorxiv, 2023
|
|
8U82
 
 | | KCTD5/Cullin3/Gbeta1gamma2 Complex: State B From Composite RELION Multi-body Refinement Map | | Descriptor: | BTB/POZ domain-containing protein KCTD5, Cullin-3, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Kuntz, D.A, Nguyen, D.M, Narayanan, N, Prive, G.G. | | Deposit date: | 2023-09-15 | | Release date: | 2023-10-11 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Structure and dynamics of a pentameric KCTD5/Cullin3/G beta gamma E3 ubiquitin ligase complex
Biorxiv, 2023
|
|
