6DAV
 
 | | Crystal Structure of Human DHFR complexed with NADP and N10formyltetrahydrofolate | | Descriptor: | 1,2-ETHANEDIOL, Dihydrofolate reductase, N-(4-{[(2,4-diaminopteridin-6-yl)methyl](hydroxymethyl)amino}benzene-1-carbonyl)-L-glutamic acid, ... | | Authors: | Mayclin, S.J, Dranow, D.M, Lorimer, D.D. | | Deposit date: | 2018-05-02 | | Release date: | 2018-05-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal Structure of Human DHFR complexed with NADP and N10formyltetrahydrofolate
to be published
|
|
7JWL
 
 | | Crystal Structure of Pseudomonas aeruginosa Penicillin Binding Protein 3 (PAE-PBP3) bound to ETX0462 | | Descriptor: | CHLORIDE ION, ETX0462 (Bound form), Peptidoglycan D,D-transpeptidase FtsI | | Authors: | Mayclin, S.J, Abendroth, J, Horanyi, P.S, Sylvester, M, Wu, X, Shapiro, A, Moussa, S, Durand-Reville, T.F. | | Deposit date: | 2020-08-25 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Rational design of a new antibiotic class for drug-resistant infections.
Nature, 597, 2021
|
|
6UGX
 
 | | Crystal structure of the Fc fragment of PF06438179/GP1111 an infliximab biosimilar in a primative orthorhombic crystal form, Lot A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Mayclin, S.J, Edwards, T.E. | | Deposit date: | 2019-09-26 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of PF-06438179/GP1111, an Infliximab Biosimilar.
BioDrugs, 34, 2020
|
|
6UGY
 
 | | Crystal structure of the Fc fragment of anti-TNFa antibody infliximab (Remicade) in a primative orthorhombic crystal form, Lot C | | Descriptor: | ACETATE ION, Remicade Fc, ZINC ION, ... | | Authors: | Mayclin, S.J, Edwards, T.E. | | Deposit date: | 2019-09-26 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of PF-06438179/GP1111, an Infliximab Biosimilar.
BioDrugs, 34, 2020
|
|
6UGW
 
 | | Crystal structure of the Fc fragment of PF06438179/GP1111 an infliximab biosimilar in a C-centered orthorhombic crystal form, Lot A | | Descriptor: | ACETATE ION, PF-06438179/GP1111 Fc, ZINC ION, ... | | Authors: | Mayclin, S.J, Edwards, T.E. | | Deposit date: | 2019-09-26 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of PF-06438179/GP1111, an Infliximab Biosimilar.
BioDrugs, 34, 2020
|
|
5VH5
 
 | | Crystal Structure of Fc fragment of anti-TNFa antibody infliximab | | Descriptor: | ACETATE ION, Infliximab Fc, ZINC ION, ... | | Authors: | Mayclin, S.J, Edwards, T.E, Lerch, T.F, Conlan, H, Sharpe, P. | | Deposit date: | 2017-04-12 | | Release date: | 2017-05-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Infliximab crystal structures reveal insights into self-association.
MAbs, 9, 2017
|
|
5VH3
 
 | | Crystal structure of Fab fragment of the anti-TNFa antibody infliximab in a C-centered orthorhombic crystal form | | Descriptor: | 1,2-ETHANEDIOL, Infliximab Fab Heavy Chain, Infliximab Fab Light Chain | | Authors: | Mayclin, S.J, Edwards, T.E, Lerch, T.F, Conlan, H, Sharpe, P. | | Deposit date: | 2017-04-12 | | Release date: | 2017-05-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Infliximab crystal structures reveal insights into self-association.
MAbs, 9, 2017
|
|
5VH4
 
 | | Crystal structure of Fab fragment of anti-TNFa antibody infliximab in an I-centered orthorhombic crystal form | | Descriptor: | 1,2-ETHANEDIOL, Infliximab Fab Heavy Chain, Infliximab Fab Light Chain, ... | | Authors: | Mayclin, S.J, Edwards, T.E, Lerch, T.F, Conlan, H, Sharpe, P. | | Deposit date: | 2017-04-12 | | Release date: | 2017-05-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Infliximab crystal structures reveal insights into self-association.
MAbs, 9, 2017
|
|
8GBU
 
 | | Hepatitis B capsid Y132A mutant with compound AB-506 | | Descriptor: | (1-methyl-1H-1,2,4-triazol-3-yl)methyl {(1S)-4-[(3-chloro-4-fluorophenyl)carbamoyl]-7-fluoro-2,3-dihydro-1H-inden-1-yl}carbamate, 1,2-ETHANEDIOL, Capsid protein | | Authors: | Horanyi, P.S, Mayclin, S.J. | | Deposit date: | 2023-02-28 | | Release date: | 2023-09-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Design, synthesis, and structure-activity relationship of a bicyclic HBV capsid assembly modulator chemotype leading to the identification of clinical candidate AB-506.
Bioorg.Med.Chem.Lett., 94, 2023
|
|
6UGU
 
 | | Crystal structure of the Fab fragment of anti-TNFa antibody infliximab (Remicade) in a C-centered orthorhombic crystal form, Lot C | | Descriptor: | PF06438179 Fab Heavy Chain, PF06438179 Fab Light Chain | | Authors: | Lerch, T.F, Sharpe, P, Mayclin, S.J, Edwards, T.E, Polleck, S, Rouse, J.C, Conlan, H.D. | | Deposit date: | 2019-09-26 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structures of PF-06438179/GP1111, an Infliximab Biosimilar.
BioDrugs, 34, 2020
|
|
6UGS
 
 | | Crystal structure of the Fab fragment of PF06438179/GP1111 an infliximab biosimilar in a C-centered orthorhombic crystal form, Lot A | | Descriptor: | Infliximab (Remicade) Fab Heavy Chain, Infliximab (Remicade) Fab Light Chain | | Authors: | Lerch, T.F, Sharpe, P, Mayclin, S.J, Edwards, T.E, Polleck, S, Rouse, J.C, Conlan, H. | | Deposit date: | 2019-09-26 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structures of PF-06438179/GP1111, an Infliximab Biosimilar.
BioDrugs, 34, 2020
|
|
6UGT
 
 | | Crystal structure of the Fab fragment of PF06438179/GP1111 an infliximab biosimilar in a I-centered orthorhombic crystal form, Lot A | | Descriptor: | 1,2-ETHANEDIOL, PF06438179 Fab Heavy Chain, PF06438179 Fab Light Chain | | Authors: | Lerch, T.F, Sharpe, P, Mayclin, S.J, Edwards, T.E, Polleck, S, Rouse, J.C, Conlan, H.D. | | Deposit date: | 2019-09-26 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structures of PF-06438179/GP1111, an Infliximab Biosimilar.
BioDrugs, 34, 2020
|
|
6UGV
 
 | | Crystal structure of the Fab fragment of anti-TNFa antibody infliximab (Remicade) in a I-centered orthorhombic crystal form, Lot C | | Descriptor: | 1,2-ETHANEDIOL, Infliximab Fab Heavy Chain, Infliximab Fab Light Chain, ... | | Authors: | Lerch, T.F, Sharpe, P, Mayclin, S.J, Edwards, T.E, Polleck, S, Rouse, J.C, Conlan, H.D. | | Deposit date: | 2019-09-26 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of PF-06438179/GP1111, an Infliximab Biosimilar.
BioDrugs, 34, 2020
|
|
6X9F
 
 | | Pseudomonas aeruginosa MurC with AZ8074 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Horanyi, P.S, Mayclin, S.J, Durand-Reville, T.F, Lorimer, D.D, Edwards, T.E, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2020-06-02 | | Release date: | 2020-09-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Pseudomonas aeruginosa MurC with AZ8074
To Be Published
|
|
6X9N
 
 | | Pseudomonas aeruginosa MurC with AZ5595 | | Descriptor: | (2R)-2-({4-[(5-tert-butyl-1-methyl-1H-pyrazol-3-yl)amino]-1H-pyrazolo[3,4-d]pyrimidin-6-yl}amino)-2-phenylethan-1-ol, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Horanyi, P.S, Mayclin, S.J, Durand-Reville, T.F, Lorimer, D.D, Edwards, T.E, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2020-06-03 | | Release date: | 2020-09-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Pseudomonas aeruginosa MurC with AZ5595
To Be Published
|
|
5IXM
 
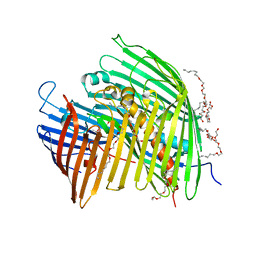 | | The LPS Transporter LptDE from Yersinia pestis, core complex | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, LPS-assembly lipoprotein LptE, LPS-assembly protein LptD | | Authors: | Botos, I, Mayclin, S.J, McCarthy, J.G, Buchanan, S.K. | | Deposit date: | 2016-03-23 | | Release date: | 2016-05-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.746 Å) | | Cite: | Structural and Functional Characterization of the LPS Transporter LptDE from Gram-Negative Pathogens.
Structure, 24, 2016
|
|
8DV0
 
 | |
5SCO
 
 | |
5SD3
 
 | |
5SCV
 
 | |
5SCR
 
 | |
7RZO
 
 | |
7RZM
 
 | |
5K9F
 
 | |
5KOB
 
 | |
