3NAD
 
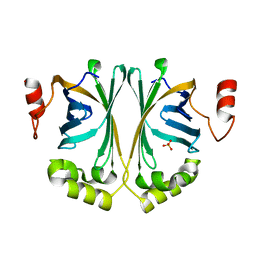 | | Crystal Structure of Phenolic Acid Decarboxylase from Bacillus pumilus UI-670 | | Descriptor: | Ferulate decarboxylase, SULFATE ION | | Authors: | Matte, A, Grosse, S, Bergeron, H, Abokitse, K, Lau, P.C.K. | | Deposit date: | 2010-06-01 | | Release date: | 2010-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural analysis of Bacillus pumilus phenolic acid decarboxylase, a lipocalin-fold enzyme.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
1OEN
 
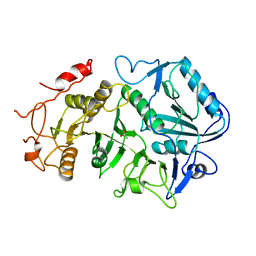 | | PHOSPHOENOLPYRUVATE CARBOXYKINASE | | Descriptor: | ACETATE ION, PHOSPHOENOLPYRUVATE CARBOXYKINASE | | Authors: | Matte, A, Goldie, H, Sweet, R.M, Delbaere, L.T.J. | | Deposit date: | 1995-09-08 | | Release date: | 1996-11-08 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Escherichia coli phosphoenolpyruvate carboxykinase: a new structural family with the P-loop nucleoside triphosphate hydrolase fold.
J.Mol.Biol., 256, 1996
|
|
1DBO
 
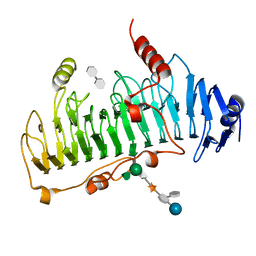 | | CRYSTAL STRUCTURE OF CHONDROITINASE B | | Descriptor: | 4-deoxy-alpha-D-glucopyranose-(1-3)-[beta-D-glucopyranose-(1-4)]2-O-methyl-beta-L-fucopyranose-(1-4)-beta-D-xylopyranose-(1-4)-alpha-D-glucopyranuronic acid-(1-2)-[alpha-L-rhamnopyranose-(1-4)]alpha-D-mannopyranose, 4-deoxy-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose, CHONDROITINASE B | | Authors: | Huang, W, Matte, A, Li, Y, Kim, Y.S, Linhardt, R.J, Su, H, Cygler, M. | | Deposit date: | 1999-11-03 | | Release date: | 2000-01-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of chondroitinase B from Flavobacterium heparinum and its complex with a disaccharide product at 1.7 A resolution.
J.Mol.Biol., 294, 1999
|
|
1AQ2
 
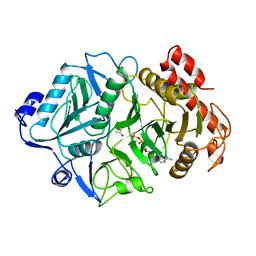 | | PHOSPHOENOLPYRUVATE CARBOXYKINASE | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Tari, L.W, Matte, A, Goldie, H, Delbaere, L.T.J. | | Deposit date: | 1997-08-05 | | Release date: | 1998-10-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mg(2+)-Mn2+ clusters in enzyme-catalyzed phosphoryl-transfer reactions.
Nat.Struct.Biol., 4, 1997
|
|
3B8M
 
 | |
3B8N
 
 | |
3B8O
 
 | |
3BE5
 
 | | Crystal structure of FitE (crystal form 1), a group III periplasmic siderophore binding protein | | Descriptor: | CHLORIDE ION, Putative iron compound-binding protein of ABC transporter family | | Authors: | Shi, R, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2007-11-16 | | Release date: | 2008-10-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Trapping open and closed forms of FitE-A group III periplasmic binding protein.
Proteins, 75, 2008
|
|
2I6R
 
 | | Crystal structure of E. coli HypE, a hydrogenase maturation protein | | Descriptor: | HypE protein | | Authors: | Rangarajan, E.S, Proteau, A, Iannuzzi, P, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2006-08-29 | | Release date: | 2007-10-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structure of [NiFe] hydrogenase maturation protein HypE from Escherichia coli and its interaction with HypF.
J.Bacteriol., 190, 2008
|
|
3LA0
 
 | | Crystal Structure of UreE from Helicobacter pylori (metal of unknown identity bound) | | Descriptor: | UNKNOWN ATOM OR ION, Urease accessory protein ureE | | Authors: | Shi, R, Munger, C, Assinas, A, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2010-01-06 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Crystal Structures of Apo and Metal-Bound Forms of the UreE Protein from Helicobacter pylori: Role of Multiple Metal Binding Sites
Biochemistry, 49, 2010
|
|
3L9Z
 
 | | Crystal Structure of UreE from Helicobacter pylori (apo form) | | Descriptor: | Urease accessory protein ureE | | Authors: | Shi, R, Munger, C, Assinas, A, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2010-01-06 | | Release date: | 2010-08-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Crystal Structures of Apo and Metal-Bound Forms of the UreE Protein from Helicobacter pylori: Role of Multiple Metal Binding Sites
Biochemistry, 49, 2010
|
|
3HBN
 
 | | Crystal structure PseG-UDP complex from Campylobacter jejuni | | Descriptor: | CHLORIDE ION, GLYCEROL, UDP-sugar hydrolase, ... | | Authors: | Rangarajan, E.S, Proteau, A, Cygler, M, Matte, A, Sulea, T, Schoenhofen, I.C. | | Deposit date: | 2009-05-04 | | Release date: | 2009-05-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and functional analysis of Campylobacter jejuni PseG: a udp-sugar hydrolase from the pseudaminic acid biosynthetic pathway.
J.Biol.Chem., 284, 2009
|
|
3NY0
 
 | | Crystal Structure of UreE from Helicobacter pylori (Ni2+ bound form) | | Descriptor: | NICKEL (II) ION, Urease accessory protein ureE | | Authors: | Shi, R, Munger, C, Assinas, A, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2010-07-14 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Crystal Structures of Apo and Metal-Bound Forms of the UreE Protein from Helicobacter pylori: Role of Multiple Metal Binding Sites
Biochemistry, 49, 2010
|
|
3NXZ
 
 | | Crystal Structure of UreE from Helicobacter pylori (Cu2+ bound form) | | Descriptor: | COPPER (II) ION, Urease accessory protein ureE | | Authors: | Shi, R, Munger, C, Assinas, A, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2010-07-14 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structures of Apo and Metal-Bound Forms of the UreE Protein from Helicobacter pylori: Role of Multiple Metal Binding Sites
Biochemistry, 49, 2010
|
|
1FC4
 
 | | 2-AMINO-3-KETOBUTYRATE COA LIGASE | | Descriptor: | 2-AMINO-3-KETOBUTYRATE CONENZYME A LIGASE, 2-AMINO-3-KETOBUTYRIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Schmidt, A, Matte, A, Li, Y, Sivaraman, J, Larocque, R, Schrag, J.D, Smith, C, Sauve, V, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2000-07-17 | | Release date: | 2001-05-02 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional structure of 2-amino-3-ketobutyrate CoA ligase from Escherichia coli complexed with a PLP-substrate intermediate: inferred reaction mechanism.
Biochemistry, 40, 2001
|
|
3LVL
 
 | | Crystal Structure of E.coli IscS-IscU complex | | Descriptor: | Cysteine desulfurase, NifU-like protein, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Shi, R, Proteau, A, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2010-02-22 | | Release date: | 2010-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for Fe-S cluster assembly and tRNA thiolation mediated by IscS protein-protein interactions.
Plos Biol., 8, 2010
|
|
3LVJ
 
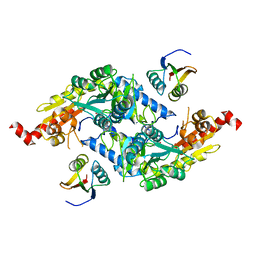 | | Crystal Structure of E.coli IscS-TusA complex (form 1) | | Descriptor: | Cysteine desulfurase, PYRIDOXAL-5'-PHOSPHATE, Sulfurtransferase tusA | | Authors: | Shi, R, Proteau, A, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2010-02-22 | | Release date: | 2010-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.435 Å) | | Cite: | Structural basis for Fe-S cluster assembly and tRNA thiolation mediated by IscS protein-protein interactions.
Plos Biol., 8, 2010
|
|
3LVK
 
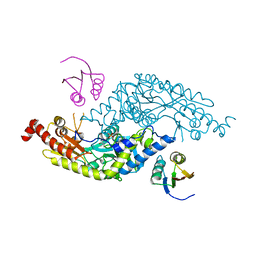 | | Crystal Structure of E.coli IscS-TusA complex (form 2) | | Descriptor: | Cysteine desulfurase, PYRIDOXAL-5'-PHOSPHATE, Sulfurtransferase tusA | | Authors: | Shi, R, Proteau, A, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2010-02-22 | | Release date: | 2010-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.442 Å) | | Cite: | Structural basis for Fe-S cluster assembly and tRNA thiolation mediated by IscS protein-protein interactions.
Plos Biol., 8, 2010
|
|
3LVM
 
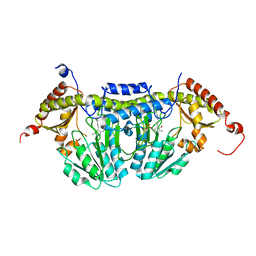 | | Crystal Structure of E.coli IscS | | Descriptor: | Cysteine desulfurase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Shi, R, Proteau, A, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2010-02-22 | | Release date: | 2010-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for Fe-S cluster assembly and tRNA thiolation mediated by IscS protein-protein interactions.
Plos Biol., 8, 2010
|
|
2RB9
 
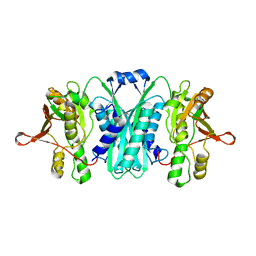 | | Crystal structure of E.coli HypE | | Descriptor: | HypE protein | | Authors: | Asinas, A.E, Rangarajan, E.S, Min, T, Matte, A, Proteau, A, Munger, C, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2007-09-18 | | Release date: | 2007-10-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of [NiFe] hydrogenase maturation protein HypE from Escherichia coli and its interaction with HypF.
J.Bacteriol., 190, 2008
|
|
1P9N
 
 | | Crystal structure of Escherichia coli MobB. | | Descriptor: | Molybdopterin-guanine dinucleotide biosynthesis protein B, SULFATE ION | | Authors: | Rangarajan, S.E, Tocilj, A, Li, Y, Iannuzzi, P, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2003-05-12 | | Release date: | 2003-05-20 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecules of Escherichia coli MobB assemble into densely packed hollow cylinders in a crystal lattice with 75% solvent content.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1SBZ
 
 | | Crystal Structure of dodecameric FMN-dependent Ubix-like Decarboxylase from Escherichia coli O157:H7 | | Descriptor: | FLAVIN MONONUCLEOTIDE, Probable aromatic acid decarboxylase | | Authors: | Rangarajan, E.S, Li, Y, Iannuzzi, P, Tocilj, A, Hung, L.-W, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2004-02-11 | | Release date: | 2004-10-26 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a dodecameric FMN-dependent UbiX-like decarboxylase (Pad1) from Escherichia coli O157: H7.
Protein Sci., 13, 2004
|
|
2VHE
 
 | | PglD-CoA complex: An acetyl transferase from Campylobacter jejuni | | Descriptor: | ACETYLTRANSFERASE, COENZYME A, SULFATE ION | | Authors: | Rangarajan, E.S, Ruane, K.M, Sulea, T, Watson, D.C, Proteau, A, Leclerc, S, Cygler, M, Matte, A, Young, N.M. | | Deposit date: | 2007-11-21 | | Release date: | 2008-01-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and Active Site Residues of Pgld, an N-Acetyltransferase from the Bacillosamine Synthetic Pathway Required for N-Glycan Synthesis in Campylobacter Jejuni
Biochemistry, 47, 2008
|
|
2GML
 
 | | Crystal Structure of Catalytic Domain of E.coli RluF | | Descriptor: | Ribosomal large subunit pseudouridine synthase F | | Authors: | Sunita, S, Zhenxing, H, Swaathi, J, Cygler, M, Matte, A, Sivaraman, J. | | Deposit date: | 2006-04-06 | | Release date: | 2006-07-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Domain Organization and Crystal Structure of the Catalytic Domain of E.coli RluF, a Pseudouridine Synthase that Acts on 23S rRNA
J.Mol.Biol., 359, 2006
|
|
2HXW
 
 | | Crystal Structure of Peb3 from Campylobacter jejuni | | Descriptor: | CITRATE ANION, Major antigenic peptide PEB3 | | Authors: | Rangarajan, E.S, Bhatia, S, Watson, D.C, Munger, C, Cygler, M, Matte, A, Young, N.M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2006-08-04 | | Release date: | 2007-05-01 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural context for protein N-glycosylation in bacteria: The structure of PEB3, an adhesin from Campylobacter jejuni.
Protein Sci., 16, 2007
|
|
