1DEJ
 
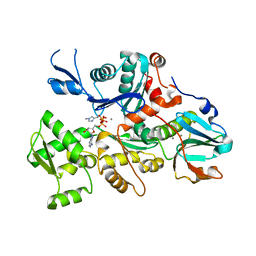 | | CRYSTAL STRUCTURE OF A DICTYOSTELIUM/TETRAHYMENA CHIMERA ACTIN (MUTANT 646: Q228K/T229A/A230Y/A231K/S232E/E360H) IN COMPLEX WITH HUMAN GELSOLIN SEGMENT 1 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, CHIMERIC ACTIN, ... | | Authors: | Matsuura, Y, Stewart, M, Kawamoto, M, Kamiya, N, Saeki, K, Yasunaga, T, Wakabayashi, T. | | Deposit date: | 1999-11-15 | | Release date: | 2000-03-01 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for the higher Ca(2+)-activation of the regulated actin-activated myosin ATPase observed with Dictyostelium/Tetrahymena actin chimeras.
J.Mol.Biol., 296, 2000
|
|
1C0G
 
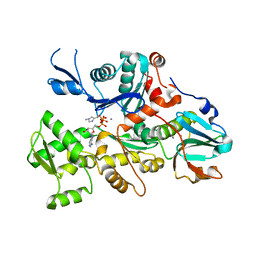 | | CRYSTAL STRUCTURE OF 1:1 COMPLEX BETWEEN GELSOLIN SEGMENT 1 AND A DICTYOSTELIUM/TETRAHYMENA CHIMERA ACTIN (MUTANT 228: Q228K/T229A/A230Y/E360H) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, PROTEIN (CHIMERIC ACTIN), ... | | Authors: | Matsuura, Y, Stewart, M, Kawamoto, M, Kamiya, N, Saeki, K, Yasunaga, T, Wakabayashi, T. | | Deposit date: | 1999-07-16 | | Release date: | 2000-03-01 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the higher Ca(2+)-activation of the regulated actin-activated myosin ATPase observed with Dictyostelium/Tetrahymena actin chimeras.
J.Mol.Biol., 296, 2000
|
|
1C0F
 
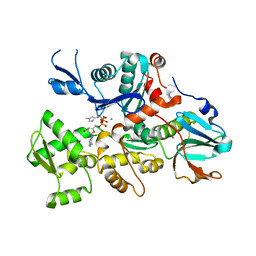 | | CRYSTAL STRUCTURE OF DICTYOSTELIUM CAATP-ACTIN IN COMPLEX WITH GELSOLIN SEGMENT 1 | | Descriptor: | ACTIN, ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, ... | | Authors: | Matsuura, Y, Stewart, M, Kawamoto, M, Kamiya, N, Saeki, K, Yasunaga, T, Wakabayashi, T. | | Deposit date: | 1999-07-16 | | Release date: | 2000-03-01 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for the higher Ca(2+)-activation of the regulated actin-activated myosin ATPase observed with Dictyostelium/Tetrahymena actin chimeras
J.Mol.Biol., 296, 2000
|
|
8HE0
 
 | |
8HE3
 
 | |
8HKW
 
 | |
6IU7
 
 | |
6IUA
 
 | |
6KC4
 
 | | Crystal structure of human Fer SH2 domain bound to a phosphopeptide (DEpYENVD) | | Descriptor: | Tyrosine-protein kinase Fer, phosphopeptide (EDpYENVD) | | Authors: | Matsuura, Y. | | Deposit date: | 2019-06-27 | | Release date: | 2019-09-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | High-resolution structural analysis shows how different crystallographic environments can induce alternative modes of binding of a phosphotyrosine peptide to the SH2 domain of Fer tyrosine kinase.
Protein Sci., 28, 2019
|
|
1UN0
 
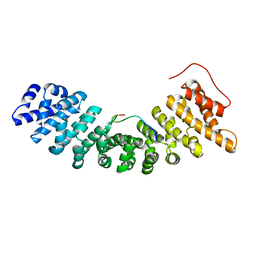 | | Crystal Structure of Yeast Karyopherin (Importin) alpha in complex with a Nup2p N-terminal fragment | | Descriptor: | IMPORTIN ALPHA SUBUNIT, NUCLEOPORIN NUP2 | | Authors: | Matsuura, Y, Lange, A, Harreman, M.T, Corbett, A.H, Stewart, M. | | Deposit date: | 2003-09-03 | | Release date: | 2003-10-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis for Nup2P Function in Cargo Release and Karyopherin Recycling in Nuclear Import
Embo J., 22, 2003
|
|
3AA9
 
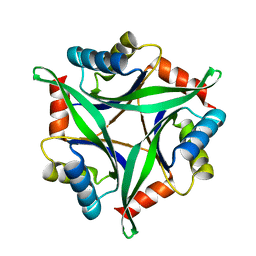 | | Crystal Structure Analysis of the Mutant CutA1 (E61V) from E. coli | | Descriptor: | Divalent-cation tolerance protein cutA | | Authors: | Matsuura, Y, Tanaka, T, Bagautdinov, B, Kunishima, N, Yutani, K. | | Deposit date: | 2009-11-12 | | Release date: | 2010-08-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Remarkable improvement in the heat stability of CutA1 from Escherichia coli by rational protein design
J.Biochem., 148, 2010
|
|
3AA8
 
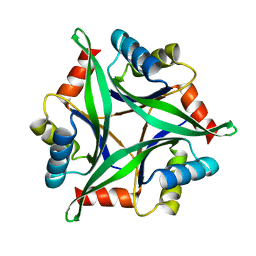 | | Crystal Structure Analysis of the Mutant CutA1 (S11V/E61V) from E. coli | | Descriptor: | Divalent-cation tolerance protein cutA | | Authors: | Matsuura, Y, Tanaka, T, Bagautdinov, B, Kunishima, N, Yutani, K. | | Deposit date: | 2009-11-12 | | Release date: | 2010-08-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Remarkable improvement in the heat stability of CutA1 from Escherichia coli by rational protein design
J.Biochem., 148, 2010
|
|
351C
 
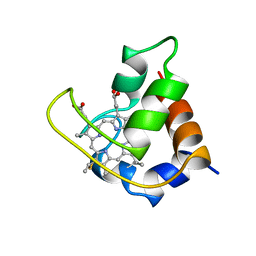 | |
3AH6
 
 | | Remarkable improvement of the heat stability of CutA1 from E.coli by rational protein designing | | Descriptor: | Divalent-cation tolerance protein cutA | | Authors: | Matsuura, Y, Tanaka, T, Bagautdinov, B, Kunishima, N, Yutani, K. | | Deposit date: | 2010-04-15 | | Release date: | 2010-08-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Remarkable improvement in the heat stability of CutA1 from Escherichia coli by rational protein design
J.Biochem., 148, 2010
|
|
451C
 
 | |
2C1T
 
 | | Structure of the Kap60p:Nup2 complex | | Descriptor: | IMPORTIN ALPHA SUBUNIT, NUCLEOPORIN NUP2 | | Authors: | Matsuura, Y, Stewart, M. | | Deposit date: | 2005-09-20 | | Release date: | 2005-11-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Nup50/Npap60 Function in Nuclear Import Complex Disassembly and Importin Recycling
Embo J., 24, 2005
|
|
2C1M
 
 | | Nup50:importin-alpha complex | | Descriptor: | IMPORTIN-ALPHA2 SUBUNIT, NUCLEOPORIN 50 KDA | | Authors: | Matsuura, Y, Stewart, M. | | Deposit date: | 2005-09-16 | | Release date: | 2005-11-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Nup50/Npap60 Function in Nuclear Protein Import Complex Disassembly and Importin Recycling.
Embo J., 24, 2005
|
|
2TAA
 
 | | STRUCTURE AND POSSIBLE CATALYTIC RESIDUES OF TAKA-AMYLASE A | | Descriptor: | CALCIUM ION, TAKA-AMYLASE A | | Authors: | Kusunoki, M, Matsuura, Y, Tanaka, N, Kakudo, M. | | Deposit date: | 1982-10-18 | | Release date: | 1982-10-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure and possible catalytic residues of Taka-amylase A
J.Biochem.(Tokyo), 95, 1984
|
|
1GCY
 
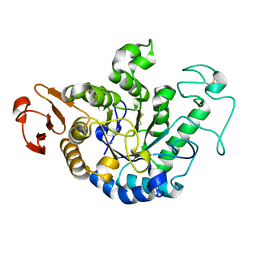 | | HIGH RESOLUTION CRYSTAL STRUCTURE OF MALTOTETRAOSE-FORMING EXO-AMYLASE | | Descriptor: | CALCIUM ION, GLUCAN 1,4-ALPHA-MALTOTETRAHYDROLASE | | Authors: | Mezaki, Y, Katsuya, Y, Kubota, M, Matsuura, Y. | | Deposit date: | 2000-08-14 | | Release date: | 2000-08-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallization and structural analysis of intact maltotetraose-forming exo-amylase from Pseudomonas stutzeri.
Biosci.Biotechnol.Biochem., 65, 2001
|
|
1FLE
 
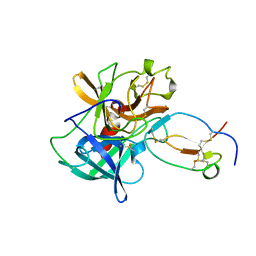 | | CRYSTAL STRUCTURE OF ELAFIN COMPLEXED WITH PORCINE PANCREATIC ELASTASE | | Descriptor: | ELAFIN, ELASTASE | | Authors: | Tsunemi, M, Matsuura, Y, Sakakibara, S, Katsube, Y. | | Deposit date: | 1996-07-04 | | Release date: | 1997-01-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of an elastase-specific inhibitor elafin complexed with porcine pancreatic elastase determined at 1.9 A resolution.
Biochemistry, 35, 1996
|
|
1QPK
 
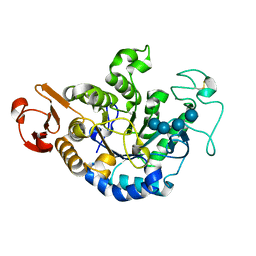 | | MUTANT (D193G) MALTOTETRAOSE-FORMING EXO-AMYLASE IN COMPLEX WITH MALTOTETRAOSE | | Descriptor: | CALCIUM ION, PROTEIN (MALTOTETRAOSE-FORMING AMYLASE), alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Yoshioka, Y, Hasegawa, K, Matsuura, Y, Katsube, Y, Kubota, M. | | Deposit date: | 1999-05-26 | | Release date: | 1999-11-17 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Roles of catalytic residues in alpha-amylases as evidenced by the structures of the product-complexed mutants of a maltotetraose-forming amylase.
Protein Eng., 12, 1999
|
|
1QI3
 
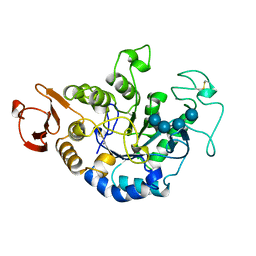 | | MUTANT (D193N) MALTOTETRAOSE-FORMING EXO-AMYLASE IN COMPLEX WITH MALTOTETRAOSE | | Descriptor: | CALCIUM ION, PROTEIN (EXO-MALTOTETRAOHYDROLASE), alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Hasegawa, K, Kubota, M, Matsuura, Y. | | Deposit date: | 1999-06-01 | | Release date: | 1999-11-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Roles of catalytic residues in alpha-amylases as evidenced by the structures of the product-complexed mutants of a maltotetraose-forming amylase.
Protein Eng., 12, 1999
|
|
1QI5
 
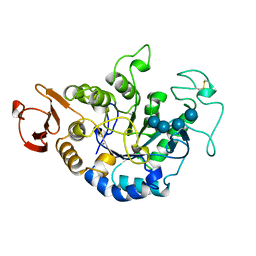 | | MUTANT (D294N) MALTOTETRAOSE-FORMING EXO-AMYLASE IN COMPLEX WITH MALTOTETRAOSE | | Descriptor: | CALCIUM ION, PROTEIN (EXO-MALTOTETRAOHYDROLASE), alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Hasegawa, K, Kubota, M, Matsuura, Y. | | Deposit date: | 1999-06-03 | | Release date: | 1999-11-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Roles of catalytic residues in alpha-amylases as evidenced by the structures of the product-complexed mutants of a maltotetraose-forming amylase.
Protein Eng., 12, 1999
|
|
1QI4
 
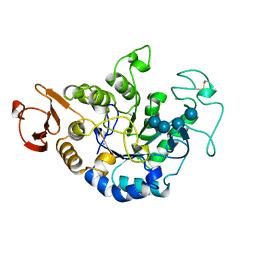 | | MUTANT (E219G) MALTOTETRAOSE-FORMING EXO-AMYLASE IN COMPLEX WITH MALTOTETRAOSE | | Descriptor: | CALCIUM ION, PROTEIN (EXO-MALTOTETRAOHYDROLASE), alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Hasegawa, K, Kubota, M, Matsuura, Y. | | Deposit date: | 1999-06-01 | | Release date: | 1999-11-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Roles of catalytic residues in alpha-amylases as evidenced by the structures of the product-complexed mutants of a maltotetraose-forming amylase.
Protein Eng., 12, 1999
|
|
1BF2
 
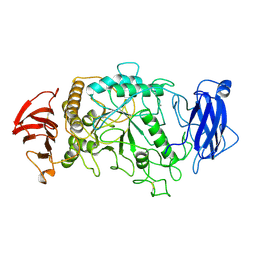 | | STRUCTURE OF PSEUDOMONAS ISOAMYLASE | | Descriptor: | CALCIUM ION, ISOAMYLASE | | Authors: | Katsuya, Y, Mezaki, Y, Kubota, M, Matsuura, Y. | | Deposit date: | 1998-05-26 | | Release date: | 1998-08-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional structure of Pseudomonas isoamylase at 2.2 A resolution.
J.Mol.Biol., 281, 1998
|
|
