1QB4
 
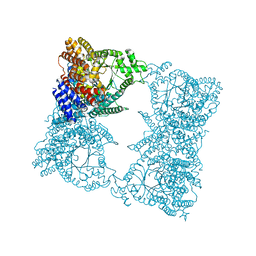 | | CRYSTAL STRUCTURE OF MN(2+)-BOUND PHOSPHOENOLPYRUVATE CARBOXYLASE | | Descriptor: | ASPARTIC ACID, MANGANESE (II) ION, PHOSPHOENOLPYRUVATE CARBOXYLASE | | Authors: | Matsumura, H, Terada, M, Shirakata, S, Inoue, T, Yoshinaga, T, Izui, K, Kai, Y. | | Deposit date: | 1999-04-30 | | Release date: | 2002-05-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Plausible phosphoenolpyruvate binding site revealed by 2.6 A structure of Mn2+-bound phosphoenolpyruvate carboxylase from Escherichia coli
FEBS Lett., 458, 1999
|
|
6KYJ
 
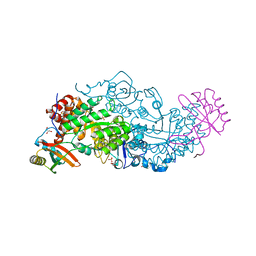 | | Hybrid-Rubisco (rice RbcL and sorghum RbcS) in complex with sulfate ions | | Descriptor: | GLYCEROL, Ribulose bisphosphate carboxylase large chain, Ribulose bisphosphate carboxylase small chain, ... | | Authors: | Matsumura, H, Yoshizawa, T, Tanaka, S, Yoshikawa, H. | | Deposit date: | 2019-09-19 | | Release date: | 2020-09-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Hybrid Rubisco with Complete Replacement of Rice Rubisco Small Subunits by Sorghum Counterparts Confers C 4 Plant-like High Catalytic Activity.
Mol Plant, 13, 2020
|
|
6KYI
 
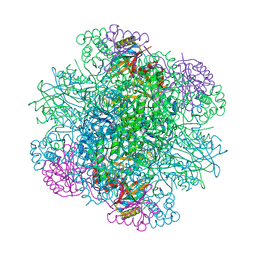 | | Rice Rubisco in complex with sulfate ions | | Descriptor: | GLYCEROL, Ribulose bisphosphate carboxylase large chain, Ribulose bisphosphate carboxylase small chain, ... | | Authors: | Matsumura, H, Yoshizawa, T, Tanaka, S, Yoshikawa, H. | | Deposit date: | 2019-09-19 | | Release date: | 2020-09-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Hybrid Rubisco with Complete Replacement of Rice Rubisco Small Subunits by Sorghum Counterparts Confers C 4 Plant-like High Catalytic Activity.
Mol Plant, 13, 2020
|
|
3AXM
 
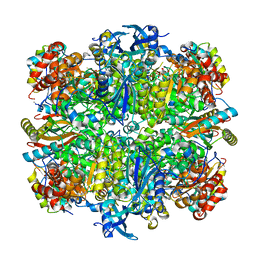 | | Structure of rice Rubisco in complex with 6PG | | Descriptor: | 6-PHOSPHOGLUCONIC ACID, MAGNESIUM ION, Ribulose bisphosphate carboxylase large chain, ... | | Authors: | Matsumura, H, Mizohata, E, Ishida, H, Kogami, A, Ueno, T, Makino, A, Inoue, T, Yokota, A, Mae, T, Kai, Y. | | Deposit date: | 2011-04-11 | | Release date: | 2012-04-11 | | Last modified: | 2013-06-05 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of rice Rubisco and implications for activation induced by positive effectors NADPH and 6-phosphogluconate
J.Mol.Biol., 422, 2012
|
|
3AXK
 
 | | Structure of rice Rubisco in complex with NADP(H) | | Descriptor: | GLYCEROL, MAGNESIUM ION, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Matsumura, H, Mizohata, E, Ishida, H, Kogami, A, Ueno, T, Makino, A, Inoue, T, Yokota, A, Mae, T, Kai, Y. | | Deposit date: | 2011-04-11 | | Release date: | 2012-04-11 | | Last modified: | 2013-06-05 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of rice Rubisco and implications for activation induced by positive effectors NADPH and 6-phosphogluconate
J.Mol.Biol., 422, 2012
|
|
3B1K
 
 | |
3B1J
 
 | | Crystal structure of Glyceraldehyde-3-Phosphate Dehydrogenase complexed with CP12 in the presence of copper from Synechococcus elongatus | | Descriptor: | COPPER (II) ION, CP12, Glyceraldehyde 3-phosphate dehydrogenase (NADP+), ... | | Authors: | Matsumura, H, Kai, A, Inoue, T. | | Deposit date: | 2011-07-04 | | Release date: | 2012-01-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure Basis for the Regulation of Glyceraldehyde-3-Phosphate Dehydrogenase Activity via the Intrinsically Disordered Protein CP12.
Structure, 19, 2011
|
|
3B20
 
 | | Crystal structure of Glyceraldehyde-3-Phosphate Dehydrogenase complexed with NADfrom Synechococcus elongatus" | | Descriptor: | Glyceraldehyde 3-phosphate dehydrogenase (NADP+), NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | Authors: | Matsumura, H, Kai, A, Maeda, T, Inoue, T. | | Deposit date: | 2011-07-17 | | Release date: | 2012-01-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.398 Å) | | Cite: | Structure Basis for the Regulation of Glyceraldehyde-3-Phosphate Dehydrogenase Activity via the Intrinsically Disordered Protein CP12.
Structure, 19, 2011
|
|
2CW7
 
 | | Crystal structure of intein homing endonuclease II | | Descriptor: | Endonuclease PI-PkoII, SULFATE ION | | Authors: | Matsumura, H, Takahashi, H, Inoue, T, Hashimoto, H, Nishioka, M, Fujiwara, S, Takagi, M, Imanaka, T, Kai, Y. | | Deposit date: | 2005-06-17 | | Release date: | 2006-04-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of intein homing endonuclease II encoded in DNA polymerase gene from hyperthermophilic archaeon Thermococcus kodakaraensis strain KOD1
Proteins, 63, 2006
|
|
2CW8
 
 | | Crystal structure of intein homing endonuclease II | | Descriptor: | Endonuclease PI-PkoII, GLYCEROL, SULFATE ION | | Authors: | Matsumura, H, Takahashi, H, Inoue, T, Hashimoto, H, Nishioka, M, Fujiwara, S, Takagi, M, Imanaka, T, Kai, Y. | | Deposit date: | 2005-06-17 | | Release date: | 2006-04-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of intein homing endonuclease II encoded in DNA polymerase gene from hyperthermophilic archaeon Thermococcus kodakaraensis strain KOD1
Proteins, 63, 2006
|
|
1JQO
 
 | |
1JQN
 
 | |
2Z5G
 
 | | Crystal structure of T1 lipase F16L mutant | | Descriptor: | CALCIUM ION, CHLORIDE ION, Thermostable lipase, ... | | Authors: | Matsumura, H, Yamamoto, T, Inoue, T, Kai, Y. | | Deposit date: | 2007-07-08 | | Release date: | 2007-10-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Novel cation-pi interaction revealed by crystal structure of thermoalkalophilic lipase
Proteins, 70, 2007
|
|
2DSN
 
 | | Crystal structure of T1 lipase | | Descriptor: | CALCIUM ION, CHLORIDE ION, SODIUM ION, ... | | Authors: | Matsumura, H, Kai, Y. | | Deposit date: | 2006-07-03 | | Release date: | 2007-07-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Novel cation-pi interaction revealed by crystal structure of thermoalkalophilic lipase
Proteins, 70, 2007
|
|
3VP8
 
 | | Crystal structure of the N-terminal domain of the yeast general corepressor Tup1p | | Descriptor: | General transcriptional corepressor TUP1 | | Authors: | Matsumura, H, Kusaka, N, Nakamura, T, Tanaka, N, Sagegami, K, Uegaki, K, Inoue, T, Mukai, Y. | | Deposit date: | 2012-02-28 | | Release date: | 2012-06-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystal structure of the N-terminal domain of the yeast general corepressor Tup1p and its functional implications
J.Biol.Chem., 287, 2012
|
|
3VP9
 
 | | Crystal structure of the N-terminal domain of the yeast general corepressor Tup1p mutant | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, General transcriptional corepressor TUP1 | | Authors: | Matsumura, H, Kusaka, N, Nakamura, T, Tanaka, N, Sagegami, K, Uegaki, K, Inoue, T, Mukai, Y. | | Deposit date: | 2012-02-28 | | Release date: | 2012-06-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Crystal structure of the N-terminal domain of the yeast general corepressor Tup1p and its functional implications
J.Biol.Chem., 287, 2012
|
|
1MHO
 
 | | THE 2.0 A STRUCTURE OF HOLO S100B FROM BOVINE BRAIN | | Descriptor: | CALCIUM ION, S-100 PROTEIN | | Authors: | Matsumura, H, Shiba, T, Inoue, T, Harada, S, Yasushi, K.A.I. | | Deposit date: | 1997-09-11 | | Release date: | 1998-11-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A novel mode of target recognition suggested by the 2.0 A structure of holo S100B from bovine brain.
Structure, 6, 1998
|
|
8GZW
 
 | | Klebsiella pneumoniae FtsZ complexed with monobody (P21) | | Descriptor: | Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE, Monobody | | Authors: | Matsumura, H, Yoshizawa, T, Fujita, J, Tanaka, S, Amesaka, H. | | Deposit date: | 2022-09-27 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of a FtsZ single protofilament and a double-helical tube in complex with a monobody.
Nat Commun, 14, 2023
|
|
8GZV
 
 | | Klebsiella pneumoniae FtsZ complexed with monobody (P212121) | | Descriptor: | Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE, Monobody | | Authors: | Matsumura, H, Yoshizawa, T, Fujita, J, Tanaka, S, Amesaka, H. | | Deposit date: | 2022-09-27 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of a FtsZ single protofilament and a double-helical tube in complex with a monobody.
Nat Commun, 14, 2023
|
|
8GZX
 
 | | Escherichia coli FtsZ complexed with monobody (P212121) | | Descriptor: | Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE, Monobody | | Authors: | Matsumura, H, Yoshizawa, T, Fujita, J, Tanaka, S, Amesaka, H. | | Deposit date: | 2022-09-27 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structures of a FtsZ single protofilament and a double-helical tube in complex with a monobody.
Nat Commun, 14, 2023
|
|
8GZY
 
 | | Escherichia coli FtsZ complexed with monobody (P21) | | Descriptor: | Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE, Monobody | | Authors: | Matsumura, H, Yoshizawa, T, Fujita, J, Tanaka, S, Amesaka, H. | | Deposit date: | 2022-09-27 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of a FtsZ single protofilament and a double-helical tube in complex with a monobody.
Nat Commun, 14, 2023
|
|
5B3F
 
 | |
6L98
 
 | | Crystalline cast nephropathy-causing Bence-Jones protein AK: An entire immunoglobulin lambda light chain dimer | | Descriptor: | Bence-Jones protein lambda light chain AK | | Authors: | Nakagaki, T, Noguchi, K, Yohda, M, Odaka, M, Wakui, H, Matsumura, H. | | Deposit date: | 2019-11-08 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Multiple Myeloma-Associated Ig Light Chain Crystalline Cast Nephropathy.
Kidney Int Rep, 5, 2020
|
|
1WDD
 
 | | Crystal Structure of Activated Rice Rubisco Complexed with 2-Carboxyarabinitol-1,5-bisphosphate | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Mizohata, E, Matsumura, H, Ueno, T, Ishida, H, Inoue, T, Makino, A, Mae, T, Kai, Y. | | Deposit date: | 2004-05-13 | | Release date: | 2004-11-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structure of rice Rubisco and implications for activation induced by positive effectors NADPH and 6-phosphogluconate
J.Mol.Biol., 422, 2012
|
|
3WRV
 
 | |
