1O3X
 
 | | Crystal structure of human GGA1 GAT domain | | Descriptor: | ADP-ribosylation factor binding protein GGA1 | | Authors: | Shiba, T, Kawasaki, M, Takatsu, H, Nogi, T, Matsugaki, N, Igarashi, N, Suzuki, M, Kato, R, Nakayama, K, Wakatsuki, S. | | Deposit date: | 2003-05-08 | | Release date: | 2003-05-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular Mechanism of Membrane Recruitment of Gga by Arf in Lysosomal Protein Transport
Nat.Struct.Biol., 10, 2003
|
|
6I59
 
 | | Long wavelength native-SAD phasing of Sen1 helicase | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Basu, S, Olieric, V, Matsugaki, N, Kawano, Y, Takashi, T, Huang, C.Y, Leonarski, F, Yamada, Y, Vera, L, Olieric, N, Basquin, J, Wojdyla, J.A, Diederichs, K, Yamamoto, M, Bunk, O, Wang, M. | | Deposit date: | 2018-11-13 | | Release date: | 2019-03-13 | | Last modified: | 2019-06-12 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Long-wavelength native-SAD phasing: opportunities and challenges.
Iucrj, 6, 2019
|
|
6I5C
 
 | | Long wavelength native-SAD phasing of Tubulin-Stathmin-TTL complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Basu, S, Olieric, V, Matsugaki, N, Kawano, Y, Takashi, T, Huang, C.Y, Leonarski, F, Yamada, Y, Vera, L, Olieric, N, Basquin, J, Wojdyla, J.A, Diederichs, K, Yamamoto, M, Bunk, O, Wang, M. | | Deposit date: | 2018-11-13 | | Release date: | 2019-03-13 | | Last modified: | 2020-09-23 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Long-wavelength native-SAD phasing: opportunities and challenges.
Iucrj, 6, 2019
|
|
1WR6
 
 | | Crystal structure of GGA3 GAT domain in complex with ubiquitin | | Descriptor: | ADP-ribosylation factor binding protein GGA3, ubiquitin | | Authors: | Kawasaki, M, Shiba, T, Shiba, Y, Yamaguchi, Y, Matsugaki, N, Igarashi, N, Suzuki, M, Kato, R, Kato, K, Nakayama, K, Wakatsuki, S. | | Deposit date: | 2004-10-12 | | Release date: | 2005-06-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular mechanism of ubiquitin recognition by GGA3 GAT domain.
Genes Cells, 10, 2005
|
|
1IU1
 
 | | Crystal structure of human gamma1-adaptin ear domain | | Descriptor: | gamma1-adaptin | | Authors: | Nogi, T, Shiba, Y, Kawasaki, M, Shiba, T, Matsugaki, N, Igarashi, N, Suzuki, M, Kato, R, Takatsu, H, Nakayama, K, Wakatsuki, S. | | Deposit date: | 2002-02-19 | | Release date: | 2002-07-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the accessory protein recruitment by the gamma-adaptin ear domain.
Nat.Struct.Biol., 9, 2002
|
|
1J2J
 
 | | Crystal structure of GGA1 GAT N-terminal region in complex with ARF1 GTP form | | Descriptor: | ADP-ribosylation factor 1, ADP-ribosylation factor binding protein GGA1, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Shiba, T, Kawasaki, M, Takatsu, H, Nogi, T, Matsugaki, N, Igarashi, N, Suzuki, M, Kato, R, Nakayama, K, Wakatsuki, S. | | Deposit date: | 2003-01-05 | | Release date: | 2003-05-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular mechanism of membrane recruitment of GGA by ARF in lysosomal protein transport
NAT.STRUCT.BIOL., 10, 2003
|
|
1JWF
 
 | | Crystal Structure of human GGA1 VHS domain. | | Descriptor: | ADP-ribosylation factor binding protein GGA1 | | Authors: | Shiba, T, Takatsu, H, Nogi, T, Matsugaki, N, Kawasaki, M, Igarashi, N, Suzuki, M, Kato, R, Earnest, T, Nakayama, K, Wakatsuki, S. | | Deposit date: | 2001-09-04 | | Release date: | 2002-03-06 | | Last modified: | 2018-06-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for recognition of acidic-cluster dileucine sequence by GGA1.
Nature, 415, 2002
|
|
1JWG
 
 | | VHS Domain of human GGA1 complexed with cation-independent M6PR C-terminal Peptide | | Descriptor: | ADP-ribosylation factor binding protein GGA1, Cation-independent mannose-6-phosphate receptor, IODIDE ION | | Authors: | Shiba, T, Takatsu, H, Nogi, T, Matsugaki, N, Kawasaki, M, Igarashi, N, Suzuki, M, Kato, R, Earnest, T, Nakayama, K, Wakatsuki, S. | | Deposit date: | 2001-09-04 | | Release date: | 2002-03-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for recognition of acidic-cluster dileucine sequence by GGA1.
Nature, 415, 2002
|
|
1O3Y
 
 | | Crystal structure of mouse ARF1 (delta17-Q71L), GTP form | | Descriptor: | ADP-ribosylation factor 1, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION | | Authors: | Shiba, T, Kawasaki, M, Takatsu, H, Nogi, T, Matsugaki, N, Igarashi, N, Suzuki, M, Kato, R, Nakayama, K, Wakatsuki, S. | | Deposit date: | 2003-05-08 | | Release date: | 2003-05-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Molecular mechanism of membrane recruitment of GGA by ARF in lysosomal protein transport
Nat.Struct.Biol., 10, 2003
|
|
2A6E
 
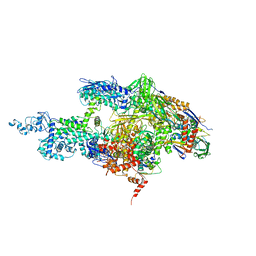 | | Crystal structure of the T. Thermophilus RNA polymerase holoenzyme | | Descriptor: | DNA-directed RNA polymerase alpha chain, DNA-directed RNA polymerase beta chain, DNA-directed RNA polymerase beta' chain, ... | | Authors: | Artsimovitch, I, Vassylyeva, M.N, Svetlov, D, Svetlov, V, Perederina, A, Igarashi, N, Matsugaki, N, Wakatsuki, S, Tahirov, T.H, Vassylyev, D.G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-02 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Allosteric modulation of the RNA polymerase catalytic reaction is an essential component of transcription control by rifamycins.
Cell(Cambridge,Mass.), 122, 2005
|
|
5D4I
 
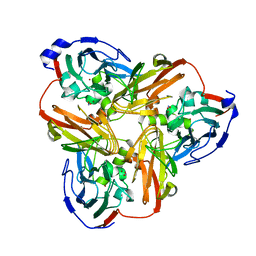 | | Intact nitrite complex of a copper nitrite reductase determined by serial femtosecond crystallography | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, NITRITE ION | | Authors: | Fukuda, Y, Tse, K.M, Nakane, T, Nakatsu, T, Suzuki, M, Sugahara, M, Inoue, S, Masuda, T, Yumoto, F, Matsugaki, N, Nango, E, Tono, K, Joti, Y, Kameshima, T, Song, C, Hatsui, T, Yabashi, M, Nureki, O, Murphy, M.E.P, Inoue, T, Iwata, S, Mizohata, E. | | Deposit date: | 2015-08-07 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Redox-coupled proton transfer mechanism in nitrite reductase revealed by femtosecond crystallography
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
2A6H
 
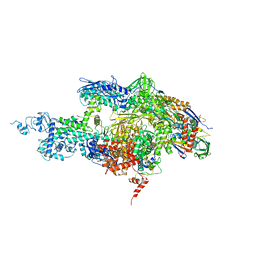 | | Crystal structure of the T. thermophilus RNA polymerase holoenzyme in complex with antibiotic sterptolydigin | | Descriptor: | DNA-directed RNA polymerase alpha chain, DNA-directed RNA polymerase beta chain, DNA-directed RNA polymerase beta' chain, ... | | Authors: | Temiakov, D, Zenkin, N, Vassylyeva, M.N, Perederina, A, Tahirov, T.H, Savkina, M, Zorov, S, Nikiforov, V, Igarashi, N, Matsugaki, N, Wakatsuki, S, Severinov, K, Vassylyev, D.G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-02 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of transcription inhibition by antibiotic streptolydigin.
Mol.Cell, 19, 2005
|
|
2A69
 
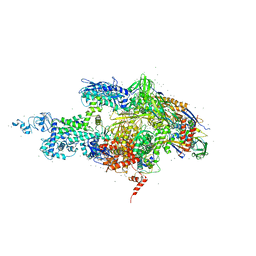 | | Crystal structure of the T. Thermophilus RNA polymerase holoenzyme in complex with antibiotic rifapentin | | Descriptor: | DNA-directed RNA polymerase alpha chain, DNA-directed RNA polymerase beta chain, DNA-directed RNA polymerase beta' chain, ... | | Authors: | Artsimovitch, I, Vassylyeva, M.N, Svetlov, D, Svetlov, V, Perederina, A, Igarashi, N, Matsugaki, N, Wakatsuki, S, Tahirov, T.H, Vassylyev, D.G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-02 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Allosteric modulation of the RNA polymerase catalytic reaction is an essential component of transcription control by rifamycins.
Cell(Cambridge,Mass.), 122, 2005
|
|
2A68
 
 | | Crystal structure of the T. thermophilus RNA polymerase holoenzyme in complex with antibiotic rifabutin | | Descriptor: | DNA-directed RNA polymerase alpha chain, DNA-directed RNA polymerase beta chain, DNA-directed RNA polymerase beta' chain, ... | | Authors: | Artsimovitch, I, Vassylyeva, M.N, Svetlov, D, Svetlov, V, Perederina, A, Igarashi, N, Matsugaki, N, Wakatsuki, S, Tahirov, T.H, Vassylyev, D.G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-01 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Allosteric modulation of the RNA polymerase catalytic reaction is an essential component of transcription control by rifamycins.
Cell(Cambridge,Mass.), 122, 2005
|
|
2BE5
 
 | | Crystal structure of the T. Thermophilus RNA polymerase holoenzyme in complex with inhibitor tagetitoxin | | Descriptor: | DNA-directed RNA polymerase alpha chain, DNA-directed RNA polymerase beta chain, DNA-directed RNA polymerase beta' chain, ... | | Authors: | Vassylyev, D.G, Svetlov, V, Vassylyeva, M.N, Perederina, A, Igarashi, N, Matsugaki, N, Wakatsuki, S, Artsimovitch, I, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-10-22 | | Release date: | 2005-11-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for transcription inhibition by tagetitoxin
Nat.Struct.Mol.Biol., 12, 2005
|
|
5D4H
 
 | | High-resolution nitrite complex of a copper nitrite reductase determined by synchrotron radiation crystallography | | Descriptor: | ACETIC ACID, COPPER (II) ION, Copper-containing nitrite reductase, ... | | Authors: | Fukuda, Y, Tse, K.M, Nakane, T, Nakatsu, T, Suzuki, M, Sugahara, M, Inoue, S, Masuda, T, Yumoto, F, Matsugaki, N, Nango, E, Tono, K, Joti, Y, Kameshima, T, Song, C, Hatsui, T, Yabashi, M, Nureki, O, Murphy, M.E.P, Inoue, T, Iwata, S, Mizohata, E. | | Deposit date: | 2015-08-07 | | Release date: | 2016-03-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Redox-coupled proton transfer mechanism in nitrite reductase revealed by femtosecond crystallography
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5D4J
 
 | | Chloride-bound form of a copper nitrite reductase from Alcaligenes faecals | | Descriptor: | ACETIC ACID, CHLORIDE ION, COPPER (II) ION, ... | | Authors: | Fukuda, Y, Tse, K.M, Nakane, T, Nakatsu, T, Suzuki, M, Sugahara, M, Inoue, S, Yumoto, F, Matsugaki, N, Nango, E, Tono, K, Joti, Y, Kameshima, T, Song, C, Yabashi, M, Nureki, O, Murphy, M.E.P, Inoue, T, Iwata, S, Mizohata, E. | | Deposit date: | 2015-08-07 | | Release date: | 2016-03-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Redox-coupled proton transfer mechanism in nitrite reductase revealed by femtosecond crystallography
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
3U3Q
 
 | | The S-SAD phased crystal structure of the ecto-domain of Death Receptor 6 (DR6) | | Descriptor: | Tumor necrosis factor receptor superfamily member 21 | | Authors: | Ru, H, Zhao, L.X, Ding, W, Jiao, L.Y, Shaw, N, Zhang, L.G, Hung, L.W, Matsugaki, N, Wakatsuki, S, Liu, Z.J. | | Deposit date: | 2011-10-06 | | Release date: | 2012-05-02 | | Last modified: | 2013-07-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | S-SAD phasing study of death receptor 6 and its solution conformation revealed by SAXS.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3U3T
 
 | | The S-SAD phased crystal structure of the ecto-domain of Death Receptor 6 (DR6) | | Descriptor: | Tumor necrosis factor receptor superfamily member 21 | | Authors: | Ru, H, Zhao, L.X, Ding, W, Jiao, L.Y, Shaw, N, Zhang, L.G, Hung, L.W, Matsugaki, N, Wakatsuki, S, Liu, Z.J. | | Deposit date: | 2011-10-06 | | Release date: | 2012-05-02 | | Last modified: | 2012-07-11 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | S-SAD phasing study of death receptor 6 and its solution conformation revealed by SAXS
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3U3S
 
 | | The S-SAD phased crystal structure of the ecto-domain of Death Receptor 6 (DR6) | | Descriptor: | Tumor necrosis factor receptor superfamily member 21 | | Authors: | Ru, H, Zhao, L.X, Ding, W, Jiao, L.Y, Shaw, N, Zhang, L.G, Hung, L.W, Matsugaki, N, Wakatsuki, S, Liu, Z.J. | | Deposit date: | 2011-10-06 | | Release date: | 2012-05-02 | | Last modified: | 2013-07-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | S-SAD phasing study of death receptor 6 and its solution conformation revealed by SAXS.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3U3P
 
 | | The S-SAD phased crystal structure of the ecto-domain of Death Receptor 6 (DR6) | | Descriptor: | Tumor necrosis factor receptor superfamily member 21 | | Authors: | Ru, H, Zhao, L.X, Ding, W, Jiao, L.Y, Shaw, N, Zhang, L.G, Hung, L.W, Matsugaki, N, Wakatsuki, S, Liu, Z.J. | | Deposit date: | 2011-10-06 | | Release date: | 2012-05-02 | | Last modified: | 2013-07-10 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | S-SAD phasing study of death receptor 6 and its solution conformation revealed by SAXS
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3U3V
 
 | | The S-SAD phased crystal structure of the ecto-domain of Death Receptor 6 (DR6) | | Descriptor: | Tumor necrosis factor receptor superfamily member 21 | | Authors: | Ru, H, Zhao, L.X, Ding, W, Jiao, L.Y, Shaw, N, Zhang, L.G, Hung, L.W, Matsugaki, N, Wakatsuki, S, Liu, Z.J. | | Deposit date: | 2011-10-06 | | Release date: | 2012-05-02 | | Last modified: | 2012-07-11 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | S-SAD phasing study of death receptor 6 and its solution conformation revealed by SAXS
Acta Crystallogr.,Sect.D, 68, 2012
|
|
5F7A
 
 | | Nitrite complex structure of copper nitrite reductase from Alcaligenes faecalis determined at 293 K | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, NITRITE ION | | Authors: | Fukuda, Y, Tse, K.M, Nakane, T, Nakatsu, T, Suzuki, M, Sugahara, M, Inoue, S, Masuda, T, Yumoto, F, Matsugaki, N, Nango, E, Tono, K, Joti, Y, Kameshima, T, Song, C, Hatsui, T, Yabashi, M, Nureki, O, Murphy, M.E.P, Inoue, T, Iwata, S, Mizohata, E. | | Deposit date: | 2015-12-07 | | Release date: | 2016-03-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Redox-coupled proton transfer mechanism in nitrite reductase revealed by femtosecond crystallography
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5F7B
 
 | | Resting state structure of CuNiR form Alcaligenes faecalis determined at 293 K | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase | | Authors: | Fukuda, Y, Tse, K.M, Nakane, T, Nakatsu, T, Suzuki, M, Sugahara, M, Inoue, S, Masuda, T, Yumoto, F, Matsugaki, N, Nango, E, Tono, K, Joti, Y, Kameshima, T, Song, C, Hatsui, T, Yabashi, M, Nureki, O, Murphy, M.E.P, Inoue, T, Iwata, S, Mizohata, E. | | Deposit date: | 2015-12-07 | | Release date: | 2016-03-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Redox-coupled proton transfer mechanism in nitrite reductase revealed by femtosecond crystallography
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
8PXK
 
 | | Structure of NADH-DEPENDENT FERREDOXIN REDUCTASE, BPHA4, solved at wavelength 5.76 A | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Ferredoxin reductase | | Authors: | El Omari, K, Duman, R, Mykhaylyk, V, Orr, C, Senda, M, Matsugaki, N, Kawano, Y, Wagner, A. | | Deposit date: | 2023-07-23 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.77 Å) | | Cite: | Experimental phasing opportunities for macromolecular crystallography at very long wavelengths.
Commun Chem, 6, 2023
|
|
