4B9W
 
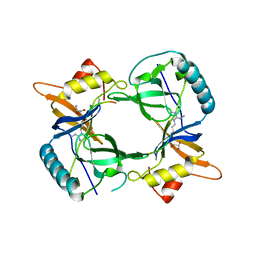 | | Structure of extended Tudor domain TD3 from mouse TDRD1 in complex with MILI peptide containing dimethylarginine 45. | | Descriptor: | GLYCEROL, PIWI-LIKE PROTEIN 2, TUDOR DOMAIN-CONTAINING PROTEIN 1 | | Authors: | Mathioudakis, N, Palencia, A, Kadlec, J, Round, A, Tripsianes, K, Sattler, M, Pillai, R.S, Cusack, S. | | Deposit date: | 2012-09-08 | | Release date: | 2012-10-17 | | Last modified: | 2019-04-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The multiple Tudor domain-containing protein TDRD1 is a molecular scaffold for mouse Piwi proteins and piRNA biogenesis factors.
Rna, 18, 2012
|
|
4B9X
 
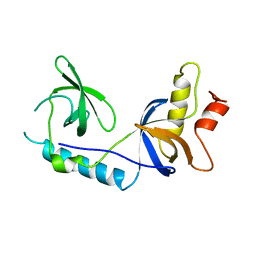 | | Structure of extended Tudor domain TD3 from mouse TDRD1 | | Descriptor: | TUDOR DOMAIN-CONTAINING PROTEIN 1 | | Authors: | Mathioudakis, N, Palencia, A, Kadlec, J, Round, A, Tripsianes, K, Sattler, M, Pillai, R.S, Cusack, S. | | Deposit date: | 2012-09-08 | | Release date: | 2012-10-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Multiple Tudor Domain-Containing Protein Tdrd1 is a Molecular Scaffold for Mouse Piwi Proteins and Pirna Biogenesis Factors.
RNA, 18, 2012
|
|
4C1Q
 
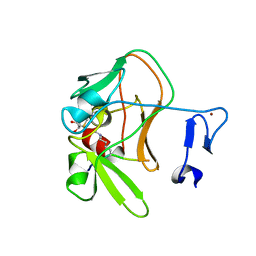 | | Crystal structure of the PRDM9 SET domain in complex with H3K4me2 and AdoHcy. | | Descriptor: | GLYCEROL, HISTONE H3.1, HISTONE-LYSINE N-METHYLTRANSFERASE PRDM9, ... | | Authors: | Mathioudakis, N, Cusack, S, Kadlec, J. | | Deposit date: | 2013-08-13 | | Release date: | 2013-10-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular Basis for the Regulation of the H3K4 Methyltransferase Activity of Prdm9.
Cell Rep., 5, 2013
|
|
5CU5
 
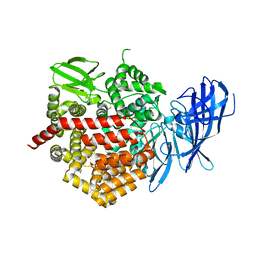 | | Crystal structure of ERAP2 without catalytic Zn(II) atom | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Endoplasmic reticulum aminopeptidase 2, ... | | Authors: | Saridakis, E, Mathioudakis, N, Giastas, P, Mavridis, I.M, Stratikos, E. | | Deposit date: | 2015-07-24 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Structural Basis for Antigenic Peptide Recognition and Processing by Endoplasmic Reticulum (ER) Aminopeptidase 2.
J.Biol.Chem., 290, 2015
|
|
5AB0
 
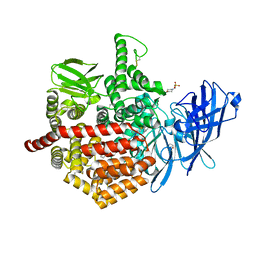 | | Crystal structure of aminopeptidase ERAP2 with ligand | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Mpakali, A, Giastas, P, Saridakis, E, Stratikos, E. | | Deposit date: | 2015-07-31 | | Release date: | 2015-09-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for Antigenic Peptide Recognition and Processing by Endoplasmic Reticulum (Er) Aminopeptidase 2.
J.Biol.Chem., 290, 2015
|
|
5AB2
 
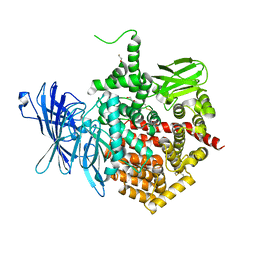 | | Crystal structure of aminopeptidase ERAP2 with ligand | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Mpakali, A, Giastas, P, Saridakis, E, Mavridis, I.M, Stratikos, E. | | Deposit date: | 2015-07-31 | | Release date: | 2015-09-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.729 Å) | | Cite: | Structural Basis for Antigenic Peptide Recognition and Processing by Endoplasmic Reticulum (Er) Aminopeptidase 2.
J.Biol.Chem., 290, 2015
|
|
