1OP9
 
 | | Complex of human lysozyme with camelid VHH HL6 antibody fragment | | Descriptor: | HL6 camel VHH fragment, Lysozyme C | | Authors: | Dumoulin, M, Last, A.M, Desmyter, A, Decanniere, K, Canet, D, Larsson, G, Spencer, A, Archer, D.B, Sasse, J, Muyldermans, S, Wyns, L, Redfield, C, Matagne, A, Robinson, C.V, Dobson, C.M. | | Deposit date: | 2003-03-05 | | Release date: | 2003-10-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | A camelid antibody fragment inhibits the formation of amyloid fibrils by human lysozyme
Nature, 424, 2003
|
|
7AG0
 
 | | Complex between the bone morphogenetic protein 2 and its antagonist Noggin | | Descriptor: | Bone morphogenetic protein 2, GLYCEROL, Noggin | | Authors: | Robert, C, Bruck, F, Herman, R, Vandevenne, M, Filee, P, Kerff, F, Matagne, A. | | Deposit date: | 2020-09-21 | | Release date: | 2022-04-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.104 Å) | | Cite: | Structural analysis of the interaction between human cytokine BMP-2 and the antagonist Noggin reveals molecular details of cell chondrogenesis inhibition.
J.Biol.Chem., 299, 2023
|
|
4I0C
 
 | | The structure of the camelid antibody cAbHuL5 in complex with human lysozyme | | Descriptor: | CHLORIDE ION, GLYCEROL, Lysozyme C, ... | | Authors: | De Genst, E, Chan, P.H, Pardon, E, Kumita, J.R, Christodoulou, J, Menzer, L, Chirgadze, D.Y, Robinson, C.V, Muyldermans, S, Matagne, A, Wyns, L, Dobson, C.M, Dumoulin, M. | | Deposit date: | 2012-11-16 | | Release date: | 2013-10-09 | | Last modified: | 2013-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A nanobody binding to non-amyloidogenic regions of the protein human lysozyme enhances partial unfolding but inhibits amyloid fibril formation.
J.Phys.Chem.B, 117, 2013
|
|
1ZMY
 
 | | cAbBCII-10 VHH framework with CDR loops of cAbLys3 grafted on it and in complex with hen egg white lysozyme | | Descriptor: | Antibody cabbcII-10:lys3, Lysozyme C | | Authors: | Saerens, D, Pellis, M, Loris, R, Pardon, E, Dumoulin, M, Matagne, A, Wyns, L, Muyldermans, S, Conrath, K. | | Deposit date: | 2005-05-11 | | Release date: | 2005-10-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Identification of a universal VHH framework to graft non-canonical antigen-binding loops of camel single-domain antibodies
J.Mol.Biol., 352, 2005
|
|
4ZV6
 
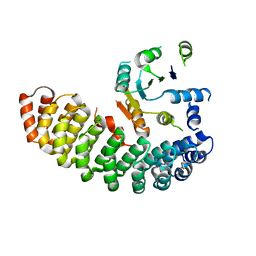 | | Crystal structure of the artificial alpharep-7 octarellinV.1 complex | | Descriptor: | AlphaRep-7, Octarellin V.1 | | Authors: | Figueroa, M, Sleutel, M, Urvoas, A, Valerio-Lepiniec, M, Minard, P, Martial, J.A, van de Weerdt, C. | | Deposit date: | 2015-05-18 | | Release date: | 2016-05-25 | | Last modified: | 2016-06-08 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | The unexpected structure of the designed protein Octarellin V.1 forms a challenge for protein structure prediction tools.
J.Struct.Biol., 195, 2016
|
|
5BOP
 
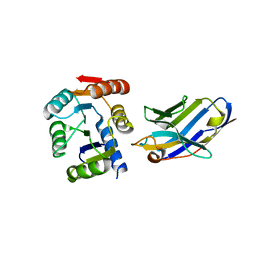 | | Crystal structure of the artificial nanobody octarellinV.1 complex | | Descriptor: | Nanobody, Octarellin V.1 | | Authors: | Figueroa, M, Sleutel, M, Pardon, E, Steyaert, J, Martial, J.A, van de Weerdt, C. | | Deposit date: | 2015-05-27 | | Release date: | 2016-05-25 | | Last modified: | 2016-06-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The unexpected structure of the designed protein Octarellin V.1 forms a challenge for protein structure prediction tools.
J.Struct.Biol., 195, 2016
|
|
3TVL
 
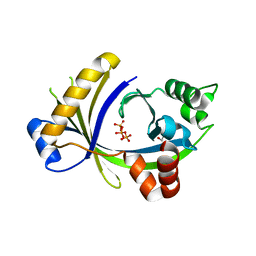 | | Complex between the human thiamine triphosphatase and triphosphate | | Descriptor: | 1,2-ETHANEDIOL, TRIPHOSPHATE, Thiamine-triphosphatase | | Authors: | Delvaux, D, Herman, R, Sauvage, E, Wins, P, Bettendorff, L, Charlier, P, Kerff, F. | | Deposit date: | 2011-09-20 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural determinants of specificity and catalytic mechanism in mammalian 25-kDa thiamine triphosphatase.
Biochim.Biophys.Acta, 1830, 2013
|
|
6FLY
 
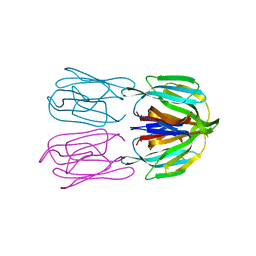 | | Structure of AcmJRL, a mannose binding jacalin related lectin from Ananas comosus, in complex with mannose. | | Descriptor: | Jacalin-like lectin, alpha-D-mannopyranose | | Authors: | Azarkan, M, Herman, R, El Mahyaoui, R, Sauvage, E, Vanden Broeck, A, Charlier, P. | | Deposit date: | 2018-01-29 | | Release date: | 2018-08-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.749 Å) | | Cite: | Biochemical and structural characterization of a mannose binding jacalin-related lectin with two-sugar binding sites from pineapple (Ananas comosus) stem.
Sci Rep, 8, 2018
|
|
6FLZ
 
 | | Structure of AcmJRL, a mannose binding jacalin related lectin from Ananas comosus, in complex with methyl-mannose. | | Descriptor: | CITRIC ACID, Jacalin-like lectin, methyl alpha-D-mannopyranoside | | Authors: | Azarkan, M, Herman, R, El Mahyaoui, R, Sauvage, E, Vanden Broeck, A, Charlier, P. | | Deposit date: | 2018-01-29 | | Release date: | 2018-08-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.895 Å) | | Cite: | Biochemical and structural characterization of a mannose binding jacalin-related lectin with two-sugar binding sites from pineapple (Ananas comosus) stem.
Sci Rep, 8, 2018
|
|
6FLW
 
 | | Structure of AcmJRL, a mannose binding jacalin related lectin from Ananas comosus. | | Descriptor: | CITRIC ACID, Jacalin-like lectin | | Authors: | Azarkan, M, Herman, R, El Mahyaoui, R, Sauvage, E, Vanden Broeck, A, Charlier, P. | | Deposit date: | 2018-01-29 | | Release date: | 2018-08-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Biochemical and structural characterization of a mannose binding jacalin-related lectin with two-sugar binding sites from pineapple (Ananas comosus) stem.
Sci Rep, 8, 2018
|
|
2WGW
 
 | | Crystal structure of the OXA-10 V117T mutant at pH 8.0 | | Descriptor: | BETA-LACTAMASE OXA-10, GLYCEROL, SULFATE ION | | Authors: | Vercheval, L, Kerff, F, Bauvois, C, Sauvage, E, Guiet, R, Charlier, P, Galleni, M. | | Deposit date: | 2009-04-27 | | Release date: | 2010-05-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Three Factors that Modulate the Activity of Class D Beta-Lactamases and Interfere with the Post-Translational Carboxylation of Lys70.
Biochem.J., 432, 2010
|
|
4M3K
 
 | | Structure of a single domain camelid antibody fragment cAb-H7S in complex with the BlaP beta-lactamase from Bacillus licheniformis | | Descriptor: | Beta-lactamase, CHLORIDE ION, Camelid heavy-chain antibody variable fragment cAb-H7S | | Authors: | Pain, C, Kerff, F, Herman, R, Sauvage, E, Preumont, S, Charlier, P, Dumoulin, M. | | Deposit date: | 2013-08-06 | | Release date: | 2014-08-06 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Probing the mechanism of aggregation of polyQ model proteins with camelid heavy-chain antibody fragments
To be Published
|
|
2WGV
 
 | | Crystal structure of the OXA-10 V117T mutant at pH 6.5 inhibited by a chloride ion | | Descriptor: | BETA-LACTAMASE OXA-10, CHLORIDE ION, CITRIC ACID, ... | | Authors: | Vercheval, L, Kerff, F, Bauvois, C, Sauvage, E, Guiet, R, Charlier, P, Galleni, M. | | Deposit date: | 2009-04-27 | | Release date: | 2010-05-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Three Factors that Modulate the Activity of Class D Beta-Lactamases and Interfere with the Post- Translational Carboxylation of Lys70.
Biochem.J., 432, 2010
|
|
4M3J
 
 | | Structure of a single-domain camelid antibody fragment cAb-H7S specific of the BlaP beta-lactamase from Bacillus licheniformis | | Descriptor: | Camelid heavy-chain antibody variable fragment cAb-H7S, SULFATE ION | | Authors: | Pain, C, Kerff, F, Herman, R, Sauvage, E, Preumont, S, Charlier, P, Dumoulin, M. | | Deposit date: | 2013-08-06 | | Release date: | 2014-08-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Probing the mechanism of aggregation of polyQ model proteins with camelid heavy-chain antibody fragments
To be Published
|
|
2WKI
 
 | | Crystal structure of the OXA-10 K70C mutant at pH 7.0 | | Descriptor: | 1,2-ETHANEDIOL, BETA-LACTAMASE OXA-10, GLYCEROL, ... | | Authors: | Vercheval, L, Bauvois, C, Kerff, F, Sauvage, E, Guiet, R, Charlier, P, Galleni, M. | | Deposit date: | 2009-06-11 | | Release date: | 2010-08-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Three Factors that Modulate the Activity of Class D Beta-Lactamases and Interfere with the Post-Translational Carboxylation of Lys70.
Biochem.J., 432, 2010
|
|
2WKH
 
 | | Crystal structure of the acyl-enzyme OXA-10 K70C-Ampicillin at pH 7 | | Descriptor: | (2R,4S)-2-[(R)-{[(2R)-2-amino-2-phenylacetyl]amino}(carboxy)methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, BETA-LACTAMASE OXA-10, SULFATE ION | | Authors: | Vercheval, L, Bauvois, C, Kerff, F, Sauvage, E, Guiet, R, Charlier, P, Galleni, M. | | Deposit date: | 2009-06-11 | | Release date: | 2010-08-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.791 Å) | | Cite: | Three Factors that Modulate the Activity of Class D Beta-Lactamases and Interfere with the Post-Translational Carboxylation of Lys70.
Biochem.J., 432, 2010
|
|
4N1H
 
 | | Structure of a single-domain camelid antibody fragment cAb-F11N in complex with the BlaP beta-lactamase from Bacillus licheniformis | | Descriptor: | Beta-lactamase, Camelid heavy-chain antibody variable fragment cAb-F11N | | Authors: | Pain, C, Kerff, F, Herman, R, Sauvage, E, Preumont, S, Charlier, P, Dumoulin, M. | | Deposit date: | 2013-10-04 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Probing the mechanism of aggregation of polyQ model proteins with camelid heavy-chain antibody fragments
To be Published
|
|
4N02
 
 | | Type 2 IDI from S. pneumoniae | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, Isopentenyl-diphosphate delta-isomerase, SULFATE ION | | Authors: | de Ruyck, J, Schubert, H.L, Poulter, C.D. | | Deposit date: | 2013-09-30 | | Release date: | 2014-07-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Determination of Kinetics and the Crystal Structure of a Novel Type 2 Isopentenyl Diphosphate: Dimethylallyl Diphosphate Isomerase from Streptococcus pneumoniae.
Chembiochem, 15, 2014
|
|
3GMU
 
 | | Crystal Structure of Beta-Lactamse Inhibitory Protein (BLIP) in Apo Form | | Descriptor: | AMMONIUM ION, Beta-lactamase inhibitory protein, SULFATE ION | | Authors: | Strynadka, N.C.J, Gretes, M, James, M.N.G. | | Deposit date: | 2009-03-15 | | Release date: | 2009-03-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Insights into positive and negative requirements for protein-protein interactions by crystallographic analysis of the beta-lactamase inhibitory proteins BLIP, BLIP-I, and BLP.
J.Mol.Biol., 389, 2009
|
|
