4TOQ
 
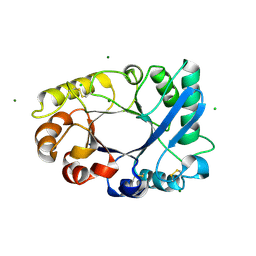 | |
5KXU
 
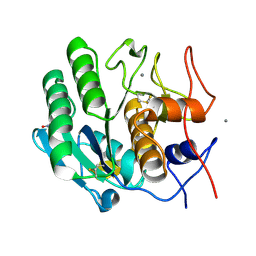 | | Structure Proteinase K determined by SACLA | | Descriptor: | CALCIUM ION, NITRATE ION, Proteinase K | | Authors: | Masuda, T, Suzuki, M, Inoue, S, Numata, K, Sugahara, M. | | Deposit date: | 2016-07-20 | | Release date: | 2017-06-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Atomic resolution structure of serine protease proteinase K at ambient temperature.
Sci Rep, 7, 2017
|
|
5KXV
 
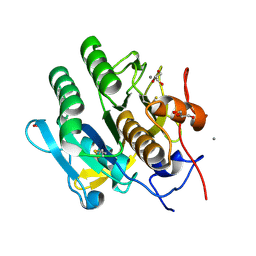 | | Structure Proteinase K at 0.98 Angstroms | | Descriptor: | CALCIUM ION, GLYCEROL, NITRATE ION, ... | | Authors: | Masuda, T, Suzuki, M, Inoue, S, Numata, K, Sugahara, M. | | Deposit date: | 2016-07-20 | | Release date: | 2017-06-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Atomic resolution structure of serine protease proteinase K at ambient temperature.
Sci Rep, 7, 2017
|
|
2RPD
 
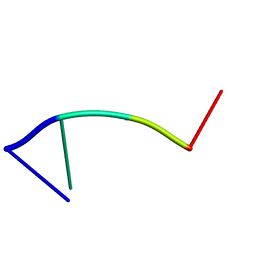 | | Mhr1p-bound ssDNA | | Descriptor: | DNA (5'-D(*DTP*DAP*DCP*DG)-3') | | Authors: | Masuda, T, Ito, Y, Shibata, T, Mikawa, T. | | Deposit date: | 2008-05-15 | | Release date: | 2009-05-26 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | A non-canonical DNA structure enables homologous recombination in various genetic systems
J.Biol.Chem., 284, 2009
|
|
2RPF
 
 | | RecO-bound ssDNA | | Descriptor: | DNA (5'-D(*DTP*DAP*DCP*DG)-3') | | Authors: | Masuda, T, Ito, Y, Shibata, T, Mikawa, T. | | Deposit date: | 2008-05-15 | | Release date: | 2009-05-26 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | A non-canonical DNA structure enables homologous recombination in various genetic systems
J.Biol.Chem., 284, 2009
|
|
2RPE
 
 | | hsRad51-bound ssDNA | | Descriptor: | DNA (5'-D(*DTP*DAP*DCP*DG)-3') | | Authors: | Masuda, T, Ito, Y, Shibata, T, Mikawa, T. | | Deposit date: | 2008-05-15 | | Release date: | 2009-05-26 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | A non-canonical DNA structure enables homologous recombination in various genetic systems
J.Biol.Chem., 284, 2009
|
|
2RPH
 
 | | RecT-bound ssDNA | | Descriptor: | DNA (5'-D(*DTP*DAP*DCP*DG)-3') | | Authors: | Masuda, T, Ito, Y, Shibata, T, Mikawa, T. | | Deposit date: | 2008-05-15 | | Release date: | 2009-05-26 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | A non-canonical DNA structure enables homologous recombination in various genetic systems
J.Biol.Chem., 284, 2009
|
|
5YYP
 
 | | Structure K137A thaumatin | | Descriptor: | GLYCEROL, L(+)-TARTARIC ACID, Preprothaumatin I | | Authors: | Masuda, T, Kigo, S, Mitsumoto, M, Ohta, K, Suzuki, M, Mikami, B, Kitabatake, N, Tani, F. | | Deposit date: | 2017-12-10 | | Release date: | 2018-03-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.01 Å) | | Cite: | Positive Charges on the Surface of Thaumatin Are Crucial for the Multi-Point Interaction with the Sweet Receptor.
Front Mol Biosci, 5, 2018
|
|
5YYQ
 
 | | Structure K78A thaumatin | | Descriptor: | GLYCEROL, L(+)-TARTARIC ACID, Preprothaumatin I | | Authors: | Masuda, T, Kigo, S, Mitsumoto, M, Ohta, K, Suzuki, M, Mikami, B, Kitabatake, N, Tani, F. | | Deposit date: | 2017-12-10 | | Release date: | 2018-03-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Positive Charges on the Surface of Thaumatin Are Crucial for the Multi-Point Interaction with the Sweet Receptor.
Front Mol Biosci, 5, 2018
|
|
5YYR
 
 | | Structure K106A thaumatin | | Descriptor: | GLYCEROL, L(+)-TARTARIC ACID, Preprothaumatin I | | Authors: | Masuda, T, Kigo, S, Ohta, K, Mitsumoto, M, Mikami, B, Suzuki, M, Kitabatake, N, Tani, F. | | Deposit date: | 2017-12-10 | | Release date: | 2018-03-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Positive Charges on the Surface of Thaumatin Are Crucial for the Multi-Point Interaction with the Sweet Receptor.
Front Mol Biosci, 5, 2018
|
|
6A4U
 
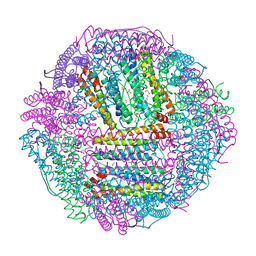 | | The first crystal structure of crustacean ferritin that is a hybrid type of H and L ferritin | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Ferritin, ... | | Authors: | Masuda, T, Mikami, B, Zang, J, Zhao, G. | | Deposit date: | 2018-06-21 | | Release date: | 2018-08-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | The first crystal structure of crustacean ferritin that is a hybrid type of H and L ferritin
Protein Sci., 27, 2018
|
|
6L8S
 
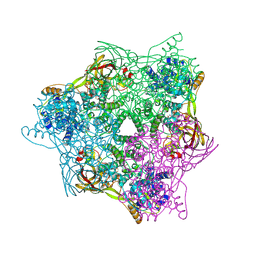 | | High resolution crystal structure of crustacean hemocyanin. | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Masuda, T, Mikami, B, Baba, S. | | Deposit date: | 2019-11-07 | | Release date: | 2020-05-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | The high-resolution crystal structure of lobster hemocyanin shows its enzymatic capability as a phenoloxidase.
Arch.Biochem.Biophys., 688, 2020
|
|
5GQP
 
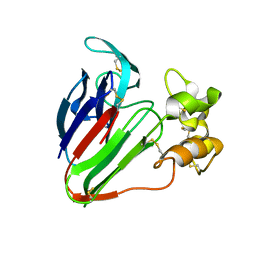 | | Thaumatin Structure at pH 8.0, orthorhombic type1 | | Descriptor: | Thaumatin I | | Authors: | Masuda, T, Sano, A, Murata, K, Okubo, K, Suzuki, M, Mikami, B. | | Deposit date: | 2016-08-08 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.296 Å) | | Cite: | Thaumatin Structure at pH 8.0, orthorhombic type1
To Be Published
|
|
5SW0
 
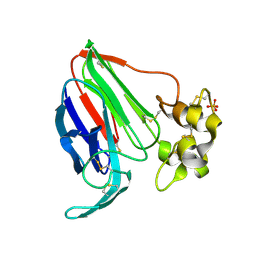 | |
5SW2
 
 | | Thaumatin Structure at pH 6.0, orthorhombic type1 | | Descriptor: | GLYCEROL, Thaumatin I | | Authors: | Masuda, T, Sano, A, Murata, K, Okubo, K, Suzuki, M, Mikami, B. | | Deposit date: | 2016-08-08 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Thaumatin Structure at pH 6.0, orthorhombic type1
To Be Published
|
|
5SW1
 
 | | Thaumatin Structure at pH 6.0 | | Descriptor: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, Thaumatin I | | Authors: | Masuda, T, Sano, A, Murata, K, Okubo, K, Suzuki, M, Mikami, B. | | Deposit date: | 2016-08-08 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Thaumatin Structure at pH 6.0
To Be Published
|
|
3A68
 
 | | Crystal structure of plant ferritin reveals a novel metal binding site that functions as a transit site for metal transfer in ferritin | | Descriptor: | ACETIC ACID, CALCIUM ION, Ferritin-4, ... | | Authors: | Masuda, T, Goto, F, Yoshihara, T, Mikami, B. | | Deposit date: | 2009-08-26 | | Release date: | 2009-12-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of plant ferritin reveals a novel metal binding site that functions as a transit site for metal transfer in ferritin
J.Biol.Chem., 285, 2010
|
|
3A9Q
 
 | | Crystal Structure Analysis of E173A variant of the soybean ferritin SFER4 | | Descriptor: | ACETIC ACID, CALCIUM ION, Ferritin-4, ... | | Authors: | Masuda, T, Goto, F, Yoshihara, T, Mikami, B. | | Deposit date: | 2009-11-05 | | Release date: | 2009-12-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.896 Å) | | Cite: | Crystal structure of plant ferritin reveals a novel metal binding site that functions as a transit site for metal transfer in ferritin
J.Biol.Chem., 285, 2010
|
|
3AJQ
 
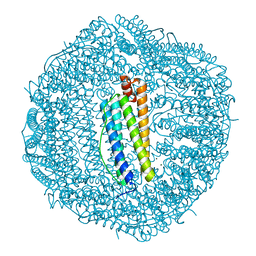 | | Crystal structure of human H ferritin E140Q mutant | | Descriptor: | Ferritin heavy chain, MAGNESIUM ION | | Authors: | Masuda, T, Mikami, B. | | Deposit date: | 2010-06-11 | | Release date: | 2010-08-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | The universal mechanism for iron translocation to the ferroxidase site in ferritin, which is mediated by the well conserved transit site
Biochem.Biophys.Res.Commun., 400, 2010
|
|
3AJP
 
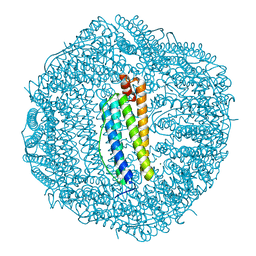 | | Crystal structure of human H ferritin E140A mutant | | Descriptor: | Ferritin heavy chain, MAGNESIUM ION | | Authors: | Masuda, T, Mikami, B. | | Deposit date: | 2010-06-11 | | Release date: | 2010-08-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | The universal mechanism for iron translocation to the ferroxidase site in ferritin, which is mediated by the well conserved transit site
Biochem.Biophys.Res.Commun., 400, 2010
|
|
3AJO
 
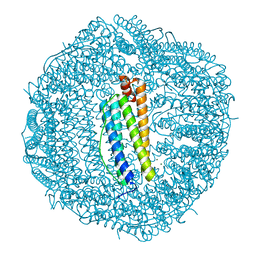 | | Crystal structure of wild-type human ferritin H chain | | Descriptor: | Ferritin heavy chain, MAGNESIUM ION | | Authors: | Masuda, T, Mikami, B. | | Deposit date: | 2010-06-11 | | Release date: | 2010-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | The universal mechanism for iron translocation to the ferroxidase site in ferritin, which is mediated by the well conserved transit site
Biochem.Biophys.Res.Commun., 400, 2010
|
|
5X9M
 
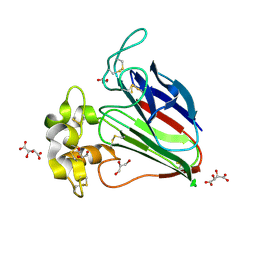 | | Structure of hyper-sweet thaumatin (D21N) | | Descriptor: | GLYCEROL, L(+)-TARTARIC ACID, Thaumatin I | | Authors: | Masuda, T, Okubo, K, Sugahara, M, Suzuki, M, Mikami, B. | | Deposit date: | 2017-03-08 | | Release date: | 2018-03-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (0.93 Å) | | Cite: | Subatomic structure of hyper-sweet thaumatin D21N mutant reveals the importance of flexible conformations for enhanced sweetness.
Biochimie, 157, 2019
|
|
5X9L
 
 | | Recombinant thaumatin I at 0.9 Angstrom | | Descriptor: | GLYCEROL, L(+)-TARTARIC ACID, Thaumatin I | | Authors: | Masuda, T, Okubo, K, Sugahara, M, Suzuki, M, Mikami, B. | | Deposit date: | 2017-03-08 | | Release date: | 2018-03-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Subatomic structure of hyper-sweet thaumatin D21N mutant reveals the importance of flexible conformations for enhanced sweetness.
Biochimie, 157, 2019
|
|
3AL7
 
 | | Recombinant thaumatin I at 1.1 A | | Descriptor: | GLYCEROL, L(+)-TARTARIC ACID, Thaumatin I | | Authors: | Masuda, T, Mikami, B, Kitabatake, N. | | Deposit date: | 2010-07-27 | | Release date: | 2011-06-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | High-resolution structure of the recombinant sweet-tasting protein thaumatin I
Acta Crystallogr.,Sect.F, 67, 2011
|
|
3ALD
 
 | | Crystal structure of sweet-tasting protein Thaumatin I at 1.10 A | | Descriptor: | GLYCEROL, L(+)-TARTARIC ACID, Thaumatin I | | Authors: | Masuda, T, Mikami, B, Kitabatake, N. | | Deposit date: | 2010-07-29 | | Release date: | 2011-06-08 | | Last modified: | 2011-11-02 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | High-resolution structure of the recombinant sweet-tasting protein thaumatin I
Acta Crystallogr.,Sect.F, 67, 2011
|
|
