1Z02
 
 | | 2-Oxoquinoline 8-Monooxygenase Component: Active site Modulation by Rieske-[2fe-2S] Center Oxidation/Reduction | | Descriptor: | 2-oxo-1,2-dihydroquinoline 8-monooxygenase, oxygenase component, FE (III) ION, ... | | Authors: | Martins, B.M, Svetlitchnaia, T, Dobbek, H. | | Deposit date: | 2005-03-01 | | Release date: | 2005-05-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 2-Oxoquinoline 8-Monooxygenase Oxygenase Component: Active Site Modulation by Rieske-[2Fe-2S] Center Oxidation/Reduction
Structure, 13, 2005
|
|
1Z03
 
 | | 2-Oxoquinoline 8-Monooxygenase Component: Active site Modulation by Rieske-[2fe-2S] Center Oxidation/Reduction | | Descriptor: | 2-oxo-1,2-dihydroquinoline 8-monooxygenase, oxygenase component, FE (III) ION, ... | | Authors: | Martins, B.M, Svetlitchnaia, T, Dobbek, H. | | Deposit date: | 2005-03-01 | | Release date: | 2005-05-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 2-Oxoquinoline 8-Monooxygenase Oxygenase Component: Active Site Modulation by Rieske-[2Fe-2S] Center Oxidation/Reduction
Structure, 13, 2005
|
|
1Z01
 
 | | 2-Oxoquinoline 8-Monooxygenase Component: Active site Modulation by Rieske-[2fe-2S] Center Oxidation/Reduction | | Descriptor: | 2-oxo-1,2-dihydroquinoline 8-monooxygenase, oxygenase component, FE (III) ION, ... | | Authors: | Martins, B.M, Svetlitchnaia, T, Dobbek, H. | | Deposit date: | 2005-03-01 | | Release date: | 2005-05-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 2-Oxoquinoline 8-Monooxygenase Oxygenase Component: Active Site Modulation by Rieske-[2Fe-2S] Center Oxidation/Reduction
Structure, 13, 2005
|
|
1U8V
 
 | | Crystal Structure of 4-Hydroxybutyryl-CoA Dehydratase from Clostridium aminobutyricum: Radical catalysis involving a [4Fe-4S] cluster and flavin | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Gamma-aminobutyrate metabolism dehydratase/isomerase, IRON/SULFUR CLUSTER | | Authors: | Martins, B.M, Dobbek, H, Cinkaya, I, Buckel, W, Messerschmidt, A. | | Deposit date: | 2004-08-07 | | Release date: | 2004-12-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of 4-hydroxybutyryl-CoA dehydratase: radical catalysis involving a [4Fe-4S] cluster and flavin.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1XDW
 
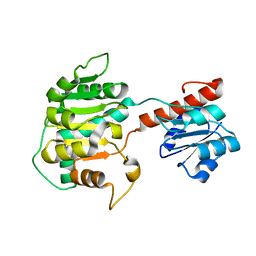 | | NAD+-dependent (R)-2-Hydroxyglutarate Dehydrogenase from Acidaminococcus fermentans | | Descriptor: | NAD+-dependent (R)-2-Hydroxyglutarate Dehydrogenase | | Authors: | Martins, B.M, Macedo-Ribeiro, S, Bresser, J, Buckel, W, Messerschmidt, A. | | Deposit date: | 2004-09-08 | | Release date: | 2005-03-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural basis for stereo-specific catalysis in NAD(+)-dependent (R)-2-hydroxyglutarate dehydrogenase from Acidaminococcus fermentans.
Febs J., 272, 2005
|
|
1J93
 
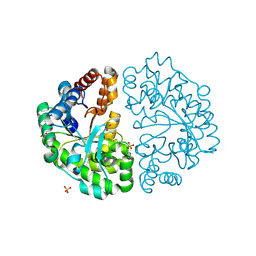 | | Crystal Structure and Substrate Binding Modeling of the Uroporphyrinogen-III Decarboxylase from Nicotiana tabacum: Implications for the Catalytic Mechanism | | Descriptor: | SULFATE ION, UROPORPHYRINOGEN DECARBOXYLASE | | Authors: | Martins, B.M, Grimm, B, Mock, H.-P, Huber, R, Messerschmidt, A. | | Deposit date: | 2001-05-23 | | Release date: | 2001-10-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure and substrate binding modeling of the uroporphyrinogen-III decarboxylase from Nicotiana tabacum. Implications for the catalytic mechanism
J.Biol.Chem., 276, 2001
|
|
2Y8N
 
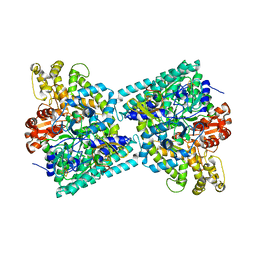 | | Crystal structure of glycyl radical enzyme | | Descriptor: | 4-HYDROXYPHENYLACETATE DECARBOXYLASE LARGE SUBUNIT, 4-HYDROXYPHENYLACETATE DECARBOXYLASE SMALL SUBUNIT, IRON/SULFUR CLUSTER | | Authors: | Martins, B.M, Blaser, M, Feliks, M, Ullmann, G.M, Selmer, T. | | Deposit date: | 2011-02-08 | | Release date: | 2011-09-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis for a Kolbe-Type Decarboxylation Catalyzed by a Glycyl Radical Enzyme.
J.Am.Chem.Soc., 133, 2011
|
|
2YAJ
 
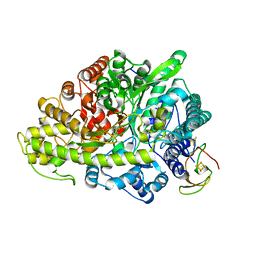 | | CRYSTAL STRUCTURE OF GLYCYL RADICAL ENZYME with bound substrate | | Descriptor: | 4-HYDROXYPHENYLACETATE, 4-HYDROXYPHENYLACETATE DECARBOXYLASE LARGE SUBUNIT, 4-HYDROXYPHENYLACETATE DECARBOXYLASE SMALL SUBUNIT, ... | | Authors: | Martins, B.M, Blaser, M, Feliks, M, Ullmann, G.M, Selmer, T. | | Deposit date: | 2011-02-23 | | Release date: | 2011-09-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.813 Å) | | Cite: | Structural Basis for a Kolbe-Type Decarboxylation Catalyzed by a Glycyl Radical Enzyme.
J.Am.Chem.Soc., 133, 2011
|
|
8CAR
 
 | | Discovery of the lanthipeptide Curvocidin and structural insights into its trifunctional synthetase CuvL | | Descriptor: | NITRATE ION, PHOSPHATE ION, Serine/threonine protein kinase | | Authors: | Martins, B.M, Sigurdsson, A, Duettmann, A.A, Jasyk, M, Dimos-Roehl, B, Schoepf, F, Gemander, M, Knittel, C.H, Schegotzki, R, Schmid, B, Kosol, S, Pommerening, L, Gonzalez-Viegas, M, Seidel, M, Huegelland, M, Leimkuehler, S, Dobbek, H, Mainz, A, Suessmuth, R. | | Deposit date: | 2023-01-24 | | Release date: | 2023-06-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Discovery of the Lanthipeptide Curvocidin and Structural Insights into its Trifunctional Synthetase CuvL.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
5NSF
 
 | | Structure of AzuAla | | Descriptor: | (2~{S})-2-azanyl-3-(2,6-dihydroazulen-1-yl)propanoic acid, CALCIUM ION, GLYCEROL, ... | | Authors: | Martins, B.M. | | Deposit date: | 2017-04-26 | | Release date: | 2019-01-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.426 Å) | | Cite: | Site-Resolved Observation of Vibrational Energy Transfer Using a Genetically Encoded Ultrafast Heater.
Angew. Chem. Int. Ed. Engl., 58, 2019
|
|
6ST5
 
 | | crystal structure of LicM2 | | Descriptor: | GLYCEROL, LicM2, MAGNESIUM ION, ... | | Authors: | Gonsior, M, Mainz, A, Hugelland, M, Kuthning, A, Tietzmann, M, Dobbek, H, Martins, B.M, Sussmuth, R. | | Deposit date: | 2019-09-10 | | Release date: | 2022-08-10 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | crystal structure of LicM2
To Be Published
|
|
3H7H
 
 | | Crystal structure of the human transcription elongation factor DSIF, hSpt4/hSpt5 (176-273) | | Descriptor: | BETA-MERCAPTOETHANOL, Transcription elongation factor SPT4, Transcription elongation factor SPT5, ... | | Authors: | Wenzel, S, Wohrl, B.M, Rosch, P, Martins, B.M. | | Deposit date: | 2009-04-27 | | Release date: | 2009-12-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of the human transcription elongation factor DSIF hSpt4 subunit in complex with the hSpt5 dimerization interface
Biochem.J., 425, 2010
|
|
5L7P
 
 | | In silico-powered specific incorporation of photocaged Dopa at multiple protein sites | | Descriptor: | (2~{S})-2-azanyl-3-[3-[(2-nitrophenyl)methoxy]-4-oxidanyl-phenyl]propanoic acid, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Hauf, M, Richter, F, Schneider, T, Martins, B.M, Baumann, T, Durkin, P, Dobbek, H, Moeglich, A, Budisa, N. | | Deposit date: | 2016-06-03 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Photoactivatable Mussel-Based Underwater Adhesive Proteins by an Expanded Genetic Code.
Chembiochem, 18, 2017
|
|
1T3Q
 
 | | Crystal structure of quinoline 2-Oxidoreductase from Pseudomonas Putida 86 | | Descriptor: | DIOXOSULFIDOMOLYBDENUM(VI) ION, FE2/S2 (INORGANIC) CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Bonin, I, Martins, B.M, Purvanov, V, Fetzner, S, Huber, R, Dobbek, H. | | Deposit date: | 2004-04-27 | | Release date: | 2004-09-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Active site geometry and substrate recognition of the molybdenum hydroxylase quinoline 2-oxidoreductase.
STRUCTURE, 12, 2004
|
|
1H98
 
 | | New Insights into Thermostability of Bacterial Ferredoxins: High Resolution Crystal Structure of the Seven-Iron Ferredoxin from Thermus thermophilus | | Descriptor: | FE3-S4 CLUSTER, FERREDOXIN, IRON/SULFUR CLUSTER | | Authors: | Macedo-Ribeiro, S, Martins, B.M, Pereira, P.J.B, Buse, G, Huber, R, Soulimane, T. | | Deposit date: | 2001-03-05 | | Release date: | 2001-11-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | New Insights Into the Thermostability of Bacterial Ferredoxins: High-Resolution Crystal Structure of the Seven-Iron Ferredoxin from Thermus Thermophilus
J.Biol.Inorg.Chem., 6, 2001
|
|
8CAV
 
 | | Discovery of the lanthipeptide Curvocidin and structural insights into its trifunctional synthetase CuvL | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CuvA, MAGNESIUM ION, ... | | Authors: | Sigurdsson, A, Martins, B.M, Duettmann, S.A, Jasyk, M, Dimos-Roehl, B, Schoepf, F, Gemannter, M, Knittel, C.H, Schnegotyzki, R, Schmid, B, Kosol, S, Gonzalez-Viegas, M, Seidel, M, Huegelland, M, Leimkuehler, S, Dobbek, H, Mainz, A, Suessmuth, R. | | Deposit date: | 2023-01-24 | | Release date: | 2023-06-14 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Discovery of the Lanthipeptide Curvocidin and Structural Insights into its Trifunctional Synthetase CuvL.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
1JDE
 
 | | K22A mutant of pyruvate, phosphate dikinase | | Descriptor: | PYRUVATE, PHOSPHATE DIKINASE, SULFATE ION | | Authors: | Ye, D, Wei, M, McGuire, M, Huang, K, Kapadia, G, Herzberg, O, Martin, B.M, Dunaway-Mariano, D. | | Deposit date: | 2001-06-13 | | Release date: | 2001-11-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Investigation of the catalytic site within the ATP-grasp domain of Clostridium symbiosum pyruvate phosphate dikinase.
J.Biol.Chem., 276, 2001
|
|
1C55
 
 | |
1C56
 
 | |
4AQ4
 
 | | substrate bound sn-glycerol-3-phosphate binding periplasmic protein ugpB from Escherichia coli | | Descriptor: | CADMIUM ION, COBALT (II) ION, GLYCEROL, ... | | Authors: | Wuttge, S, Bommer, M, Jaeger, F, Martins, B.M, Jacob, S, Licht, A, Scheffel, F, Dobbek, H, Schneider, E. | | Deposit date: | 2012-04-13 | | Release date: | 2012-10-17 | | Last modified: | 2019-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Determinants of Substrate Specificity and Biochemical Properties of the Sn-Glycerol-3-Phosphate ATP Binding Cassette Transporter (Ugpb-Aec(2) ) of Escherichia Coli.
Mol.Microbiol., 86, 2012
|
|
2FT6
 
 | |
2FT8
 
 | |
2FT7
 
 | |
2FTA
 
 | |
2OYY
 
 | | HTHP: a hexameric tyrosine-coordinated heme protein | | Descriptor: | Hexameric cytochrome, IODIDE ION, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Dobbek, H. | | Deposit date: | 2007-02-23 | | Release date: | 2007-04-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | HTHP: A Novel Class of Hexameric, Tyrosine-coordinated Heme Proteins
J.Mol.Biol., 368, 2007
|
|
