2X4Z
 
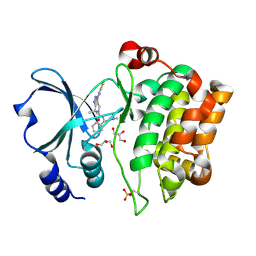 | | Crystal Structure of the Human p21-Activated Kinase 4 in Complex with PF-03758309 | | Descriptor: | GLYCEROL, PF-3758309, SERINE/THREONINE-PROTEIN KINASE PAK 4 | | Authors: | Knighton, D.R, Deng, Y, Murray, B, Guo, C, Piraino, J, Westwick, J, Zhang, C, Lamerdin, J, Dagostino, E, Loi, C.-M, Zager, M, Kraynov, E, Christensen, J, Martinez, R, Kephart, S, Marakovits, J, Karlicek, S, Bergqvist, S, Smeal, T. | | Deposit date: | 2010-02-03 | | Release date: | 2010-05-19 | | Last modified: | 2019-01-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Small-Molecule P21-Activated Kinase Inhibitor Pf- 3758309 is a Potent Inhibitor of Oncogenic Signaling and Tumor Growth.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3GB8
 
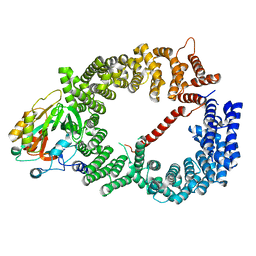 | | Crystal structure of CRM1/Snurportin-1 complex | | Descriptor: | Exportin-1, Snurportin-1 | | Authors: | Dong, X, Biswas, A, Suel, K.E, Jackson, L.K, Martinez, R, Gu, H, Chook, Y.M. | | Deposit date: | 2009-02-19 | | Release date: | 2009-03-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for leucine-rich nuclear export signal recognition by CRM1.
Nature, 458, 2009
|
|
6NZV
 
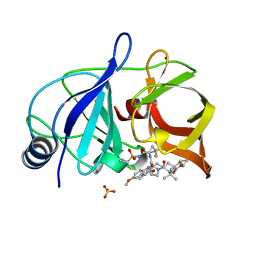 | | Crystal structure of HCV NS3/4A protease in complex with compound 12 | | Descriptor: | (1aR,5S,8S,9S,10R,22aR)-5-tert-butyl-N-[(1R,2R)-2-(difluoromethyl)-1-{[(1-methylcyclopropyl)sulfonyl]carbamoyl}cyclopropyl]-9-ethyl-14-methoxy-3,6-dioxo-1,1a,3,4,5,6,9,10,18,19,20,21,22,22a-tetradecahydro-8H-7,10-methanocyclopropa[18,19][1,10,3,6]dioxadiazacyclononadecino[11,12-b]quinoxaline-8-carboxamide, HCV NS3/4A protease, SULFATE ION, ... | | Authors: | Appleby, T.C, Taylor, J.G. | | Deposit date: | 2019-02-14 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery of the pan-genotypic hepatitis C virus NS3/4A protease inhibitor voxilaprevir (GS-9857): A component of Vosevi®.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
6NZT
 
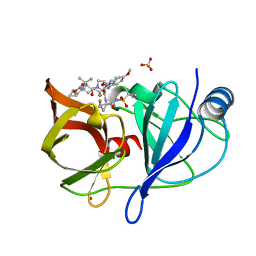 | |
6MOA
 
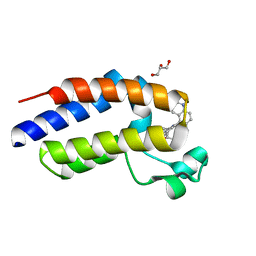 | | C-terminal bromodomain of human BRD2 in complex with 4-(2-cyclopropyl-7-(6-methylquinolin-5-yl)-1H-benzo[d]imidazol-5-yl)-3,5-dimethylisoxazole inhibitor | | Descriptor: | 4-(2-cyclopropyl-7-(6-methylquinolin-5-yl)-1H-benzo[d]imidazol-5-yl)-3,5-dimethylisoxazole, Bromodomain-containing protein 2, GLYCEROL | | Authors: | Lansdon, E.B, Newby, Z.E.R. | | Deposit date: | 2018-10-04 | | Release date: | 2019-01-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.271 Å) | | Cite: | Structure-guided discovery of a novel, potent, and orally bioavailable 3,5-dimethylisoxazole aryl-benzimidazole BET bromodomain inhibitor.
Bioorg. Med. Chem., 27, 2019
|
|
6MO7
 
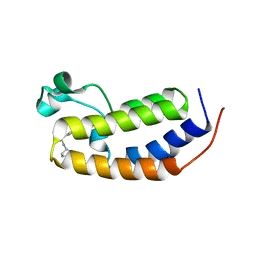 | | N-terminal bromodomain of human BRD2 with N-((4-(3-(N-cyclopentylsulfamoyl)-4-methylphenyl)-3-methylisoxazol-5-yl)methyl)acetamide inhibitor | | Descriptor: | Bromodomain-containing protein 2, N-({4-[3-(cyclopentylsulfamoyl)-4-methylphenyl]-3-methyl-1,2-oxazol-5-yl}methyl)acetamide | | Authors: | Lansdon, E.B, Newby, Z.E.R. | | Deposit date: | 2018-10-04 | | Release date: | 2019-01-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-guided discovery of a novel, potent, and orally bioavailable 3,5-dimethylisoxazole aryl-benzimidazole BET bromodomain inhibitor.
Bioorg. Med. Chem., 27, 2019
|
|
6MO9
 
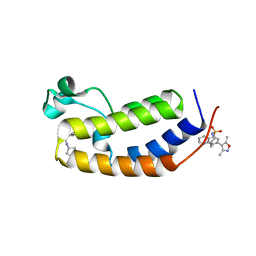 | | N-terminal bromodomain of human BRD2 in complex with N-cyclopentyl-7-(3,5-dimethylisoxazol-4-yl)quinoline-5-sulfonamide inhibitor | | Descriptor: | Bromodomain-containing protein 2, N-cyclopentyl-7-(3,5-dimethyl-1,2-oxazol-4-yl)quinoline-5-sulfonamide | | Authors: | Lansdon, E.B, Newby, Z.E.R. | | Deposit date: | 2018-10-04 | | Release date: | 2019-01-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structure-guided discovery of a novel, potent, and orally bioavailable 3,5-dimethylisoxazole aryl-benzimidazole BET bromodomain inhibitor.
Bioorg. Med. Chem., 27, 2019
|
|
6MO8
 
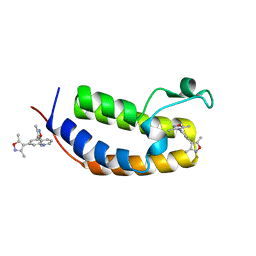 | | N-terminal bromodomain of human BRD2 in complex with 4,4'-(quinoline-5,7-diyl)bis(3,5-dimethylisoxazole) inhibitor | | Descriptor: | 5,7-bis(3,5-dimethyl-1,2-oxazol-4-yl)quinoline, Bromodomain-containing protein 2, SULFATE ION | | Authors: | Lansdon, E.B, Newby, Z.E.R. | | Deposit date: | 2018-10-04 | | Release date: | 2019-01-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-guided discovery of a novel, potent, and orally bioavailable 3,5-dimethylisoxazole aryl-benzimidazole BET bromodomain inhibitor.
Bioorg. Med. Chem., 27, 2019
|
|
6G5S
 
 | | Solution structure of the TPR domain of the cell division coordinator, CpoB | | Descriptor: | Cell division coordinator CpoB | | Authors: | Simorre, J.P, Maya Martinez, R.C, Bougault, C, Vollmer, W, Egan, A. | | Deposit date: | 2018-03-29 | | Release date: | 2018-08-08 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Induced conformational changes activate the peptidoglycan synthase PBP1B.
Mol. Microbiol., 110, 2018
|
|
6FZK
 
 | | NMR structure of UB2H, regulatory domain of PBP1b from E. coli | | Descriptor: | Penicillin-binding protein 1B | | Authors: | Simorre, J.P, Maya Martinez, R.C, Bougault, C, Egan, A.J.F, Vollmer, W. | | Deposit date: | 2018-03-15 | | Release date: | 2019-02-20 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Induced conformational changes activate the peptidoglycan synthase PBP1B.
Mol. Microbiol., 110, 2018
|
|
6G5R
 
 | |
2MKW
 
 | |
2MMX
 
 | |
6DZ8
 
 | |
