7QLF
 
 | |
1YIU
 
 | | Itch E3 ubiquitin ligase WW3 domain | | Descriptor: | Itchy E3 ubiquitin protein ligase | | Authors: | Shaw, A.Z, Martin-Malpartida, P, Morales, B, Yraola, F, Royo, M, Macias, M.J. | | Deposit date: | 2005-01-13 | | Release date: | 2005-08-02 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Phosphorylation of either Ser16 or Thr30 does not disrupt the structure of the Itch E3 ubiquitin ligase third WW domain
Proteins, 60, 2005
|
|
2B7E
 
 | | First FF domain of Prp40 Yeast Protein | | Descriptor: | Pre-mRNA processing protein PRP40 | | Authors: | Gasch, A, Wiesner, S, Martin-Malpartida, P, Ramirez-Espain, X, Ruiz, L, Macias, M.J. | | Deposit date: | 2005-10-04 | | Release date: | 2005-11-01 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | The structure of Prp40 FF1 domain and its interaction with the crn-TPR1 motif of Clf1 gives a new insight into the binding mode of FF domains.
J.Biol.Chem., 281, 2006
|
|
6Z7O
 
 | | Crystal structure of Thioredoxin T from Drosophila melanogaster | | Descriptor: | Thioredoxin-T, ZINC ION | | Authors: | Freier, R, Aragon, E, Baginski, B, Pluta, R, Martin-Malpartida, P, Torner, C, Gonzaez, C, Macias, M. | | Deposit date: | 2020-05-31 | | Release date: | 2020-10-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structures of the germline-specific Deadhead and thioredoxin T proteins from Drosophila melanogaster reveal unique features among thioredoxins.
Iucrj, 8, 2021
|
|
2N4U
 
 | |
2N4T
 
 | |
2N4W
 
 | |
2N4V
 
 | |
2N4R
 
 | |
2N4S
 
 | |
2JUP
 
 | | FBP28WW2 domain in complex with the PPLIPPPP peptide | | Descriptor: | Formin-1, Transcription elongation regulator 1 | | Authors: | Ramirez-Espain, X, Ruiz, L, Martin-Malpartida, P, Oschkinat, H, Macias, M.J. | | Deposit date: | 2007-09-01 | | Release date: | 2007-11-06 | | Last modified: | 2020-02-19 | | Method: | SOLUTION NMR | | Cite: | Structural Characterization of a New Binding Motif and a Novel Binding Mode in Group 2 WW Domains
J.Mol.Biol., 373, 2007
|
|
2JOC
 
 | | Mouse Itch 3rd domain phosphorylated in T30 | | Descriptor: | Itchy E3 ubiquitin protein ligase | | Authors: | Macias, M.J, Shaw, A.Z, Martin-Malpartida, P, Morales, B, Ruiz, L, Ramirez-Espain, X, Yraola, F, Royo, M. | | Deposit date: | 2007-03-06 | | Release date: | 2007-04-17 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | NMR Structural Studies of the ItchWW3 Domain Reveal that Phosphorylation at T30 Inhibits the Interaction with PPxY-Containing Ligands
Structure, 15, 2007
|
|
2JO9
 
 | | Mouse Itch 3rd WW domain complex with the Epstein-Barr virus latent membrane protein 2A derived peptide EEPPPPYED | | Descriptor: | Itchy E3 ubiquitin protein ligase, Latent membrane protein 2 | | Authors: | Macias, M.J, Shaw, A.Z, Martin-Malpartida, P, Morales, B, Ruiz, L, Ramirez-Espain, X, Yraola, F, Royo, M. | | Deposit date: | 2007-03-01 | | Release date: | 2007-04-17 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | NMR Structural Studies of the ItchWW3 Domain Reveal that Phosphorylation at T30 Inhibits the Interaction with PPxY-Containing Ligands
Structure, 15, 2007
|
|
2K85
 
 | | p190-A RhoGAP FF1 domain | | Descriptor: | Glucocorticoid receptor DNA-binding factor 1 | | Authors: | Bonet, R, Ruiz, L, Martin-Malpartida, P, Macias, M. | | Deposit date: | 2008-09-02 | | Release date: | 2009-05-19 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | NMR structural studies on human p190-A RhoGAPFF1 revealed that domain phosphorylation by the PDGF-receptor alpha requires its previous unfolding.
J.Mol.Biol., 389, 2009
|
|
2RM0
 
 | | FBP28WW2 domain in complex with a PPPLIPPPP peptide | | Descriptor: | Formin-1, Transcription elongation regulator 1 | | Authors: | Ramirez-Espain, X, Ruiz, L, Martin-Malpartida, P, Oschkinat, H, Macias, M.J. | | Deposit date: | 2007-09-06 | | Release date: | 2007-11-06 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural Characterization of a New Binding Motif and a Novel Binding Mode in Group 2 WW Domains
J.Mol.Biol., 373, 2007
|
|
2RLY
 
 | | FBP28WW2 domain in complex with PTPPPLPP peptide | | Descriptor: | Formin-1, Transcription elongation regulator 1 | | Authors: | Ramirez-Espain, X, Ruiz, L, Martin-Malpartida, P, Oschkinat, H, Macias, M.J. | | Deposit date: | 2007-09-03 | | Release date: | 2007-11-06 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural Characterization of a New Binding Motif and a Novel Binding Mode in Group 2 WW Domains
J.Mol.Biol., 373, 2007
|
|
2MPT
 
 | | WW3 domain of Nedd4L in complex with its HECT domain PY motif | | Descriptor: | E3 ubiquitin-protein ligase NEDD4-like | | Authors: | Escobedo, A, Macias, M.J, Gomes, T, Aragon, E, Martin-Malpartida, P, Ruiz, L. | | Deposit date: | 2014-06-02 | | Release date: | 2014-10-22 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of the Activation and Degradation Mechanisms of the E3 Ubiquitin Ligase Nedd4L.
Structure, 22, 2014
|
|
2N1O
 
 | |
5MEZ
 
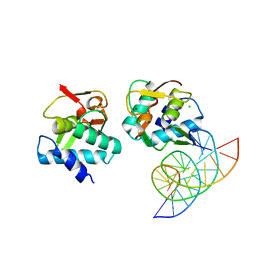 | | Crystal structure of Smad4-MH1 bound to the GGCT site. | | Descriptor: | CHLORIDE ION, DNA (5'-D(P*GP*CP*AP*GP*GP*CP*TP*AP*GP*CP*CP*TP*GP*CP*A)-3'), MH1 domain of human Smad4, ... | | Authors: | Kaczmarska, Z, Freier, R, Marquez, J.A, Macias, M.J. | | Deposit date: | 2016-11-16 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Structural basis for genome wide recognition of 5-bp GC motifs by SMAD transcription factors.
Nat Commun, 8, 2017
|
|
5MEY
 
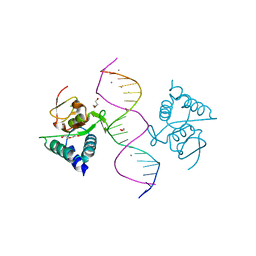 | | Crystal structure of Smad4-MH1 bound to the GGCGC site. | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kaczmarska, Z, Freier, R, Marquez, J.A, Macias, M.J. | | Deposit date: | 2016-11-16 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for genome wide recognition of 5-bp GC motifs by SMAD transcription factors.
Nat Commun, 8, 2017
|
|
5MF0
 
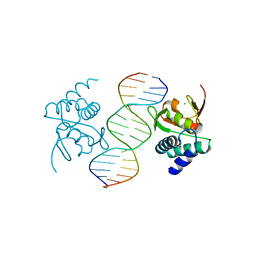 | | Crystal structure of Smad4-MH1 bound to the GGCCG site. | | Descriptor: | CHLORIDE ION, DNA (5'-D(P*AP*CP*GP*GP*GP*CP*CP*GP*CP*GP*GP*CP*CP*CP*GP*T)-3'), MH1 domain of human Smad4, ... | | Authors: | Kaczmarska, Z, Freier, R, Marquez, J.A, Macias, M.J. | | Deposit date: | 2016-11-16 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Structural basis for genome wide recognition of 5-bp GC motifs by SMAD transcription factors.
Nat Commun, 8, 2017
|
|
5NM9
 
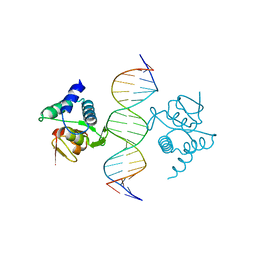 | | Crystal structure of the placozoa Trichoplax adhaerens Smad4-MH1 bound to the GGCGC site. | | Descriptor: | DNA (5'-D(P*AP*TP*GP*CP*GP*GP*GP*CP*GP*CP*GP*CP*CP*CP*GP*CP*AP*T)-3'), Mothers against decapentaplegic homolog, ZINC ION | | Authors: | Kaczmarska, Z, Freier, R, Marquez, J.A, Macias, M.J. | | Deposit date: | 2017-04-05 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structural basis for genome wide recognition of 5-bp GC motifs by SMAD transcription factors.
Nat Commun, 8, 2017
|
|
5OD6
 
 | |
5ODG
 
 | | Crystal structure of Smad3-MH1 bound to the GGCT site. | | Descriptor: | CHLORIDE ION, DNA (5'-D(P*CP*AP*GP*GP*CP*TP*AP*GP*CP*CP*TP*GP*CP*A)-3'), Mothers against decapentaplegic homolog 3, ... | | Authors: | Kaczmarska, Z, Marquez, J.A, Macias, M.J. | | Deposit date: | 2017-07-05 | | Release date: | 2017-11-15 | | Last modified: | 2017-12-27 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structural basis for genome wide recognition of 5-bp GC motifs by SMAD transcription factors.
Nat Commun, 8, 2017
|
|
6ZMU
 
 | | Crystal structure of the germline-specific thioredoxin protein Deadhead (Thioredoxin-1) from Drospohila melanogaster, P43212 | | Descriptor: | SODIUM ION, SULFATE ION, Thioredoxin-1 | | Authors: | Baginski, B, Pluta, R, Macias, M.J. | | Deposit date: | 2020-07-03 | | Release date: | 2020-10-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of the germline-specific Deadhead and thioredoxin T proteins from Drosophila melanogaster reveal unique features among thioredoxins.
Iucrj, 8, 2021
|
|
