7JLU
 
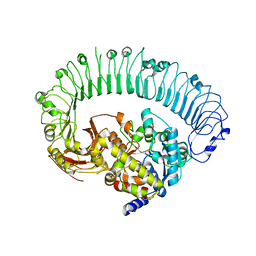 | | Structure of the activated Roq1 resistosome directly recognizing the pathogen effector XopQ | | Descriptor: | CALCIUM ION, Disease resistance protein Roq1, XopQ | | Authors: | Martin, R, Qi, T, Zhang, H, Lui, F, King, M, Toth, C, Nogales, E, Staskawicz, B.J. | | Deposit date: | 2020-07-30 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of the activated ROQ1 resistosome directly recognizing the pathogen effector XopQ.
Science, 370, 2020
|
|
7JLX
 
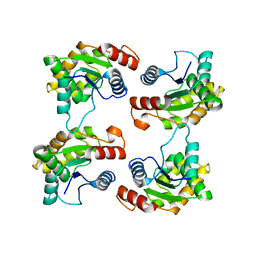 | | Structure of the activated Roq1 resistosome directly recognizing the pathogen effector XopQ (TIR domains) | | Descriptor: | Disease resistance protein Roq1 | | Authors: | Martin, R, Qi, T, Zhang, H, Lui, F, King, M, Toth, C, Nogales, E, Staskawicz, B.J. | | Deposit date: | 2020-07-30 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structure of the activated ROQ1 resistosome directly recognizing the pathogen effector XopQ.
Science, 370, 2020
|
|
7JLV
 
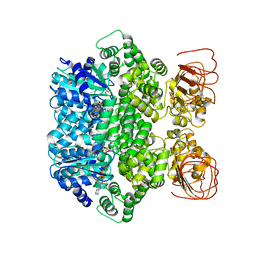 | | Structure of the activated Roq1 resistosome directly recognizing the pathogen effector XopQ | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Disease resistance protein Roq1, MAGNESIUM ION | | Authors: | Martin, R, Qi, T, Zhang, H, Lui, F, King, M, Toth, C, Nogales, E, Staskawicz, B.J. | | Deposit date: | 2020-07-30 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of the activated ROQ1 resistosome directly recognizing the pathogen effector XopQ.
Science, 370, 2020
|
|
966C
 
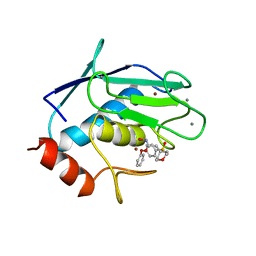 | | CRYSTAL STRUCTURE OF FIBROBLAST COLLAGENASE-1 COMPLEXED TO A DIPHENYL-ETHER SULPHONE BASED HYDROXAMIC ACID | | Descriptor: | CALCIUM ION, MMP-1, N-HYDROXY-2-[4-(4-PHENOXY-BENZENESULFONYL)-TETRAHYDRO-PYRAN-4-YL]-ACETAMIDE, ... | | Authors: | Lovejoy, B, Welch, A, Carr, S, Luong, C, Broka, C, Hendricks, R.T, Campbell, J, Walker, K, Martin, R, Van Wart, H, Browner, M.F. | | Deposit date: | 1998-08-07 | | Release date: | 1999-08-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of MMP-1 and -13 reveal the structural basis for selectivity of collagenase inhibitors.
Nat.Struct.Biol., 6, 1999
|
|
1OEC
 
 | | FGFr2 kinase domain | | Descriptor: | 4-ARYL-2-PHENYLAMINO PYRIMIDINE, FIBROBLAST GROWTH FACTOR RECEPTOR 2, SULFATE ION | | Authors: | Ceska, T.A, Owens, R, Doyle, C, Hamlyn, P, Crabbe, T, Moffat, D, Davis, J, Martin, R, Perry, M.J. | | Deposit date: | 2003-03-24 | | Release date: | 2004-10-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Crystal Structure of the Fgfr2 Tyrosine Kinase Domain in Complex with 4-Aryl-2-Phenylamino Pyrimidine Angiogenesis Inhibitors
To be Published
|
|
1GJO
 
 | | The FGFr2 tyrosine kinase domain | | Descriptor: | FIBROBLAST GROWTH FACTOR RECEPTOR 2, SULFATE ION | | Authors: | Ceska, T.A, Owens, R, Doyle, C, Hamlyn, P, Crabbe, T, Moffat, D, Davis, J, Martin, R, Perry, M.J. | | Deposit date: | 2001-07-31 | | Release date: | 2003-08-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Fgfr2 Tyrosine Kinase Domain
To be Published
|
|
1ERM
 
 | | X-RAY CRYSTAL STRUCTURE OF TEM-1 BETA LACTAMASE IN COMPLEX WITH A DESIGNED BORONIC ACID INHIBITOR (1R)-1-ACETAMIDO-2-(3-CARBOXYPHENYL)ETHANE BORONIC ACID | | Descriptor: | 1(R)-1-ACETAMIDO-2-(3-CARBOXYPHENYL)ETHYL BORONIC ACID, TEM-1 BETA-LACTAMASE | | Authors: | Ness, S, Martin, R, Kindler, A.M, Paetzel, M, Gold, M, Jones, J.B, Strynadka, N.C.J. | | Deposit date: | 2000-04-06 | | Release date: | 2000-05-10 | | Last modified: | 2012-06-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-based design guides the improved efficacy of deacylation transition state analogue inhibitors of TEM-1 beta-Lactamase(,).
Biochemistry, 39, 2000
|
|
1ZGL
 
 | | Crystal structure of 3A6 TCR bound to MBP/HLA-DR2a | | Descriptor: | HLA class II histocompatibility antigen, DR alpha chain, Myelin basic protein, ... | | Authors: | Li, Y, Huang, Y, Lue, J, Quandt, J.A, Martin, R, Mariuzza, R.A. | | Deposit date: | 2005-04-21 | | Release date: | 2005-10-18 | | Last modified: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of a human autoimmune TCR bound to a myelin basic protein self-peptide and a multiple sclerosis-associated MHC class II molecule.
Embo J., 24, 2005
|
|
2XEL
 
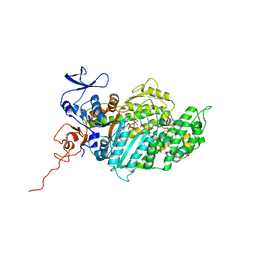 | | Molecular Mechanism of Pentachloropseudilin Mediated Inhibition of Myosin Motor Activity | | Descriptor: | 2,4-DICHLORO-6-(3,4,5-TRICHLORO-1H-PYRROL-2YL)PHENOL, ADP METAVANADATE, MAGNESIUM ION, ... | | Authors: | Chinthalapudi, K, Taft, M.H, Martin, R, Hartmann, F.K, Heissler, S.M, Tsiavaliaris, G, Gutzeit, H.O, Knoelker, H.J, Coluccio, L.M, Fedorov, R, Manstein, D.J. | | Deposit date: | 2010-05-16 | | Release date: | 2011-06-15 | | Last modified: | 2011-08-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanism and Specificity of Pentachloropseudilin-Mediated Inhibition of Myosin Motor Activity.
J.Biol.Chem., 286, 2011
|
|
5WO1
 
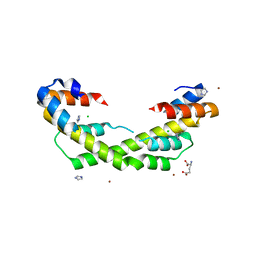 | | Chaperone Spy H96L bound to Im7 L18A L19A L37A (Im7 un-modeled) | | Descriptor: | CHLORIDE ION, GLUTAMIC ACID, IMIDAZOLE, ... | | Authors: | Horowitz, S, Koldewey, P, Martin, R, Bardwell, J.C.A. | | Deposit date: | 2017-08-01 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Visualizing chaperone-assisted protein folding.
Nat. Struct. Mol. Biol., 23, 2016
|
|
5WO3
 
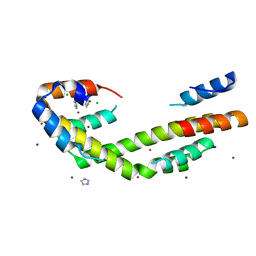 | | Chaperone Spy bound to Im7 (Im7 un-modeled) | | Descriptor: | CHLORIDE ION, IMIDAZOLE, Periplasmic chaperone Spy, ... | | Authors: | Horowitz, S, Koldewey, P, Martin, R, Bardwell, J.C.A. | | Deposit date: | 2017-08-01 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Visualizing chaperone-assisted protein folding.
Nat. Struct. Mol. Biol., 23, 2016
|
|
5WO2
 
 | | Chaperone Spy bound to Casein Fragment (Casein un-modeled) | | Descriptor: | CHLORIDE ION, IMIDAZOLE, Periplasmic chaperone Spy, ... | | Authors: | Horowitz, S, Koldewey, P, Martin, R, Bardwell, J.C.A. | | Deposit date: | 2017-08-01 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.769 Å) | | Cite: | Visualizing chaperone-assisted protein folding.
Nat. Struct. Mol. Biol., 23, 2016
|
|
5WNW
 
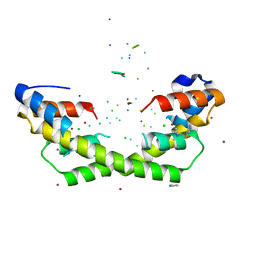 | | Chaperone Spy bound to Im7 6-45 ensemble | | Descriptor: | CHLORIDE ION, Colicin-E7 immunity protein, IMIDAZOLE, ... | | Authors: | Horowitz, S, Salmon, L, Koldewey, P, Ahlstrom, L.S, Martin, R, Xu, Q, Afonine, P.V, Trievel, R.C, Brooks, C.L, Bardwell, J.C.A. | | Deposit date: | 2017-08-01 | | Release date: | 2017-08-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Visualizing chaperone-assisted protein folding.
Nat. Struct. Mol. Biol., 23, 2016
|
|
2JHR
 
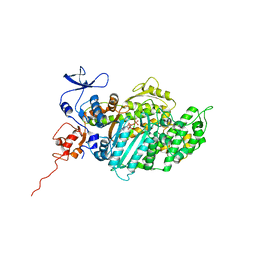 | | Crystal structure of myosin-2 motor domain in complex with ADP- metavanadate and pentabromopseudilin | | Descriptor: | ADP METAVANADATE, MAGNESIUM ION, MYOSIN-2 HEAVY CHAIN, ... | | Authors: | Fedorov, R, Boehl, M, Tsiavaliaris, G, Hartmann, F.K, Baruch, P, Brenner, B, Martin, R, Knoelker, H.J, Gutzeit, H.O, Manstein, D.J. | | Deposit date: | 2008-03-25 | | Release date: | 2009-01-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Mechanism of Pentabromopseudilin Inhibition of Myosin Motor Activity.
Nat.Struct.Mol.Biol., 16, 2009
|
|
2JJ9
 
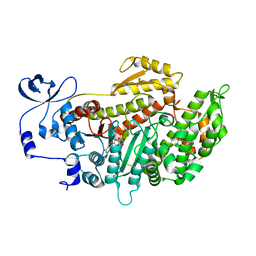 | | Crystal structure of myosin-2 in complex with ADP-metavanadate | | Descriptor: | ADP METAVANADATE, MAGNESIUM ION, MYOSIN-2 HEAVY CHAIN | | Authors: | Fedorov, R, Boehl, M, Tsiavaliaris, G, Hartmann, F.K, Baruch, P, Brenner, B, Martin, R, Knoelker, H.J, Gutzeit, H.O, Manstein, D.J. | | Deposit date: | 2008-03-25 | | Release date: | 2009-01-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Mechanism of Pentabromopseudilin Inhibition of Myosin Motor Activity.
Nat.Struct.Mol.Biol., 16, 2009
|
|
2LCQ
 
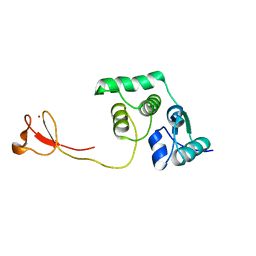 | | Solution structure of the endonuclease Nob1 from P.horikoshii | | Descriptor: | Putative toxin VapC6, ZINC ION | | Authors: | Veith, T, Martin, R, Wurm, J.P, Weis, B, Duchardt-Ferner, E, Safferthal, C, Hennig, R, Mirus, O, Bohnsack, M.T, Woehnert, J, Schleiff, E. | | Deposit date: | 2011-05-05 | | Release date: | 2011-12-14 | | Last modified: | 2012-05-09 | | Method: | SOLUTION NMR | | Cite: | Structural and functional analysis of the archaeal endonuclease Nob1.
Nucleic Acids Res., 40, 2012
|
|
1ERQ
 
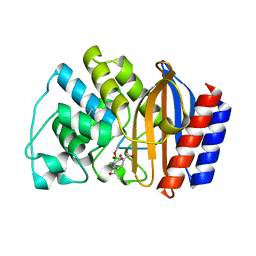 | | X-RAY CRYSTAL STRUCTURE OF TEM-1 BETA LACTAMASE IN COMPLEX WITH A DESIGNED BORONIC ACID INHIBITOR (1R)-1-ACETAMIDO-2-(3-CARBOXY-2-HYDROXYPHENYL)ETHYL BORONIC ACID | | Descriptor: | 1(R)-1-ACETAMIDO-2-(3-CARBOXY-2-HYDROXYPHENYL)ETHYL BORONIC ACID, TEM-1 BETA-LACTAMASE | | Authors: | Ness, S, Martin, R, Kindler, A.M, Paetzel, M, Gold, M, Jones, J.B, Strynadka, N.C.J. | | Deposit date: | 2000-04-06 | | Release date: | 2000-05-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-based design guides the improved efficacy of deacylation transition state analogue inhibitors of TEM-1 beta-Lactamase(,).
Biochemistry, 39, 2000
|
|
1ERO
 
 | | X-RAY CRYSTAL STRUCTURE OF TEM-1 BETA LACTAMASE IN COMPLEX WITH A DESIGNED BORONIC ACID INHIBITOR (1R)-2-PHENYLACETAMIDO-2-(3-CARBOXYPHENYL)ETHYL BORONIC ACID | | Descriptor: | (1R)-2-PHENYLACETAMIDO-2-(3-CARBOXYPHENYL)ETHYL BORONIC ACID, TEM-1 BETA-LACTAMASE | | Authors: | Ness, S, Martin, R, Kindler, A.M, Paetzel, M, Gold, M, Jones, J.B, Strynadka, N.C.J. | | Deposit date: | 2000-04-06 | | Release date: | 2000-05-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-based design guides the improved efficacy of deacylation transition state analogue inhibitors of TEM-1 beta-Lactamase(,).
Biochemistry, 39, 2000
|
|
1FV1
 
 | | STRUCTURAL BASIS FOR THE BINDING OF AN IMMUNODOMINANT PEPTIDE FROM MYELIN BASIC PROTEIN IN DIFFERENT REGISTERS BY TWO HLA-DR2 ALLELES | | Descriptor: | GLYCEROL, MAJOR HISTOCOMPATIBILITY COMPLEX ALPHA CHAIN, MAJOR HISTOCOMPATIBILITY COMPLEX BETA CHAIN, ... | | Authors: | Li, H, Mariuzza, A.R, Li, Y, Martin, R. | | Deposit date: | 2000-09-18 | | Release date: | 2000-09-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for the binding of an immunodominant peptide from myelin basic protein in different registers by two HLA-DR2 proteins.
J.Mol.Biol., 304, 2000
|
|
2XO8
 
 | | Crystal Structure of Myosin-2 in Complex with Tribromodichloropseudilin | | Descriptor: | 2,4-DICHLORO-6-(3,4,5-TRIBROMO-1H-PYRROL-2-YL)PHENOL, ADP METAVANADATE, MAGNESIUM ION, ... | | Authors: | Preller, M, Chinthalapudi, K, Martin, R, Knoelker, H.J, Manstein, D.J. | | Deposit date: | 2010-08-10 | | Release date: | 2011-05-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Inhibition of Myosin ATPase Activity by Halogenated Pseudilins: A Structure-Activity Study.
J.Med.Chem., 54, 2011
|
|
3O6F
 
 | | Crystal structure of a human autoimmune TCR MS2-3C8 bound to MHC class II self-ligand MBP/HLA-DR4 | | Descriptor: | HLA class II histocompatibility antigen, DR alpha chain, DRB1-4 beta chain, ... | | Authors: | Yin, Y, Li, Y, Martin, R, Mariuzza, R.A. | | Deposit date: | 2010-07-29 | | Release date: | 2011-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of a TCR with high affinity for self-antigen reveals basis for escape from negative selection.
Embo J., 30, 2011
|
|
830C
 
 | | COLLAGENASE-3 (MMP-13) COMPLEXED TO A SULPHONE-BASED HYDROXAMIC ACID | | Descriptor: | 4-[4-(4-CHLORO-PHENOXY)-BENZENESULFONYLMETHYL]-TETRAHYDRO-PYRAN-4-CARBOXYLIC ACID HYDROXYAMIDE, CALCIUM ION, MMP-13, ... | | Authors: | Lovejoy, B, Welch, A, Carr, S, Luong, C, Broka, C, Hendricks, R.T, Campbell, J, Walker, K, Martin, R, Van Wart, H, Browner, M.F. | | Deposit date: | 1998-08-06 | | Release date: | 1999-08-06 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of MMP-1 and -13 reveal the structural basis for selectivity of collagenase inhibitors.
Nat.Struct.Biol., 6, 1999
|
|
4NAL
 
 | | Arabidopsis thaliana IspD in complex with tribromodichloro-pseudilin | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2,4-DICHLORO-6-(3,4,5-TRIBROMO-1H-PYRROL-2-YL)PHENOL, 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase, ... | | Authors: | Kunfermann, A, Witschel, M, Illarionov, B, Martin, R, Rottmann, M, Hoffken, H.W, Seet, M, Eisenreich, W, Knolker, H.-J, Fischer, M, Bacher, A, Groll, M, Diederich, F. | | Deposit date: | 2013-10-22 | | Release date: | 2014-01-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Pseudilins: Halogenated, Allosteric Inhibitors of the Non-Mevalonate Pathway Enzyme IspD.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4NAI
 
 | | Arabidopsis thaliana IspD apo | | Descriptor: | 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase, chloroplastic, CADMIUM ION, ... | | Authors: | Kunfermann, A, Witschel, M, Illarionov, B, Martin, R, Rottmann, M, Hoffken, H.W, Seet, M, Eisenreich, W, Knolker, H.-J, Fischer, M, Bacher, A, Groll, M, Diederich, F. | | Deposit date: | 2013-10-22 | | Release date: | 2014-01-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Pseudilins: Halogenated, Allosteric Inhibitors of the Non-Mevalonate Pathway Enzyme IspD.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4NAN
 
 | | Arabidopsis thaliana IspD in complex with tetrabromo-pseudilin | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase, chloroplastic, ... | | Authors: | Kunfermann, A, Witschel, M, Illarionov, B, Martin, R, Rottmann, M, Hoffken, H.W, Seet, M, Eisenreich, W, Knolker, H.-J, Fischer, M, Bacher, A, Groll, M, Diederich, F. | | Deposit date: | 2013-10-22 | | Release date: | 2014-01-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Pseudilins: Halogenated, Allosteric Inhibitors of the Non-Mevalonate Pathway Enzyme IspD.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
