1B7B
 
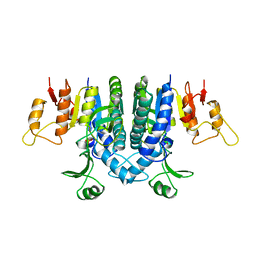 | | Carbamate kinase from Enterococcus faecalis | | Descriptor: | CARBAMATE KINASE, SULFATE ION | | Authors: | Marina, A, Alzari, P.M, Bravo, J, Uriarte, M, Barcelona, B, Fita, I, Rubio, V. | | Deposit date: | 1999-01-20 | | Release date: | 2000-01-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Carbamate kinase: New structural machinery for making carbamoyl phosphate, the common precursor of pyrimidines and arginine.
Protein Sci., 8, 1999
|
|
1ID0
 
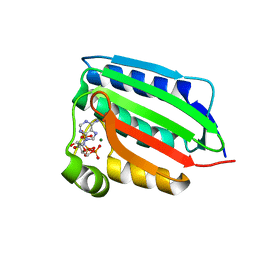 | | CRYSTAL STRUCTURE OF THE NUCLEOTIDE BOND CONFORMATION OF PHOQ KINASE DOMAIN | | Descriptor: | MAGNESIUM ION, PHOQ HISTIDINE KINASE, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Marina, A, Mott, C, Auyzenberg, A, Waldburger, C.D, Hendrickson, W.A. | | Deposit date: | 2001-04-02 | | Release date: | 2001-10-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and mutational analysis of the PhoQ histidine kinase catalytic domain. Insight into the reaction mechanism.
J.Biol.Chem., 276, 2001
|
|
2C2A
 
 | |
7PWX
 
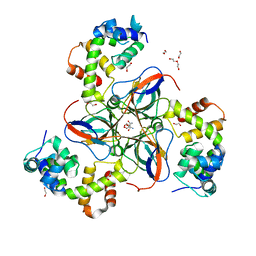 | | dUTPase from M. tuberculosis in complex with Stl | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Marina, A, Sanz-Frasquet, C. | | Deposit date: | 2021-10-07 | | Release date: | 2022-12-28 | | Last modified: | 2023-02-22 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The Bacteriophage-Phage-Inducible Chromosomal Island Arms Race Designs an Interkingdom Inhibitor of dUTPases.
Microbiol Spectr, 11, 2023
|
|
4I9E
 
 | |
4I9C
 
 | |
6S7I
 
 | |
6S7L
 
 | |
6HP3
 
 | |
6HP7
 
 | |
6HP5
 
 | | ARBITRIUM PEPTIDE RECEPTOR FROM SPBETA PHAGE | | Descriptor: | GLY-MET-PRO-ARG-GLY-ALA, SPBc2 prophage-derived uncharacterized protein YopK, SULFATE ION | | Authors: | Marina, A, Gallego del Sol, F. | | Deposit date: | 2018-09-19 | | Release date: | 2019-02-20 | | Last modified: | 2019-04-17 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Deciphering the Molecular Mechanism Underpinning Phage Arbitrium Communication Systems.
Mol.Cell, 74, 2019
|
|
8HE6
 
 | | Crystal structure of a fosfomycin and bleomycin resistant protein (ALL3014) from Anabaena/Nostoc cyanobacterium at 1.70 A resolution | | Descriptor: | All3014 protein, CALCIUM ION, MAGNESIUM ION | | Authors: | Chatterjee, A, Singh, P.K, Singh, T.P, Marina, A, Sharma, S, Rai, L.C. | | Deposit date: | 2022-11-07 | | Release date: | 2022-11-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of a fosfomycin and bleomycin resistant protein (ALL3014) from Anabaena/Nostoc cyanobacterium at 1.70 A resolution
To Be Published
|
|
3ZF0
 
 | | Phage dUTPases control transfer of virulence genes by a proto-oncogenic G protein-like mechanism. (Staphylococcus bacteriophage 80alpha dUTPase D81A mutant with dUpNHpp). | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, DUTPASE, NICKEL (II) ION | | Authors: | Tormo-Mas, M.A, Donderis, J, Garcia-Caballer, M, Alt, A, Mir-Sanchis, I, Marina, A, Penades, J.R. | | Deposit date: | 2012-12-10 | | Release date: | 2013-01-30 | | Last modified: | 2013-04-17 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Phage Dutpases Control Transfer of Virulence Genes by a Proto-Oncogenic G Protein-Like Mechanism.
Mol.Cell, 49, 2013
|
|
3GL9
 
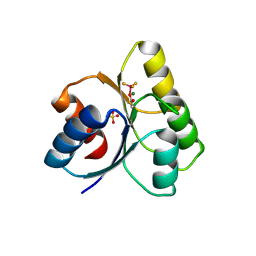 | | The structure of a histidine kinase-response regulator complex sheds light into two-component signaling and reveals a novel cis autophosphorylation mechanism | | Descriptor: | MAGNESIUM ION, Response regulator, SULFATE ION | | Authors: | Casino, P, Rubio, V, Marina, A. | | Deposit date: | 2009-03-11 | | Release date: | 2009-10-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Insight into Partner Specificity and Phosphoryl Transfer in Two-Component Signal Transduction
Cell(Cambridge,Mass.), 139, 2009
|
|
2WE4
 
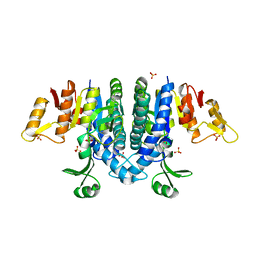 | | Carbamate kinase from Enterococcus faecalis bound to a sulfate ion and two water molecules, which mimic the substrate carbamyl phosphate | | Descriptor: | CARBAMATE KINASE 1, SULFATE ION | | Authors: | Ramon-Maiques, S, Marina, A, Gil-Ortiz, F, Rubio, V. | | Deposit date: | 2009-03-27 | | Release date: | 2010-03-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Substrate Binding and Catalysis in Carbamate Kinase Ascertained by Crystallographic and Site-Directed Mutagenesis Studies. Movements and Significance of a Unique Globular Subdomain of This Key Enzyme for Fermentative ATP Production in Bacteria.
J.Mol.Biol., 397, 2010
|
|
2WE5
 
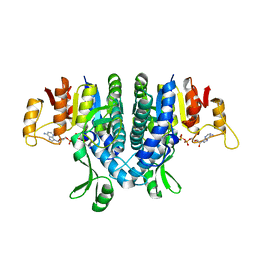 | | Carbamate kinase from Enterococcus faecalis bound to MgADP | | Descriptor: | ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, CARBAMATE KINASE 1, ... | | Authors: | Ramon-Maiques, S, Marina, A, Rubio, V. | | Deposit date: | 2009-03-27 | | Release date: | 2010-03-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Substrate Binding and Catalysis in Carbamate Kinase Ascertained by Crystallographic and Site- Directed Mutagenesis Studies. Movements and Significance of a Unique Globular Subdomain of This Key Enzyme for Fermentative ATP Production in Bacteria.
J.Mol.Biol., 397, 2010
|
|
7Q0N
 
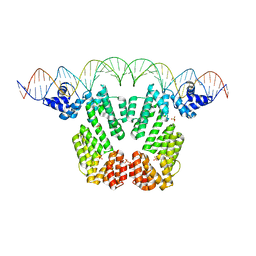 | | Arbitrium receptor from Katmira phage | | Descriptor: | Arbitrium receptor, DNA (45-MER), SULFATE ION | | Authors: | Gallego del Sol, F, Marina, A. | | Deposit date: | 2021-10-15 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insights into the mechanism of action of the arbitrium communication system in SPbeta phages.
Nat Commun, 13, 2022
|
|
7OJT
 
 | | Crystal structure of unliganded PatA, a membrane associated acyltransferase from Mycobacterium smegmatis | | Descriptor: | GLYCEROL, Phosphatidylinositol mannoside acyltransferase | | Authors: | Anso, I, Wang, L, Marina, A, Paez-Perez, E.D, Perrone, S, Lowary, T.L, Trastoy, B, Guerin, M.E. | | Deposit date: | 2021-05-17 | | Release date: | 2021-10-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.67 Å) | | Cite: | Molecular ruler mechanism and interfacial catalysis of the integral membrane acyltransferase PatA.
Sci Adv, 7, 2021
|
|
7P4A
 
 | | Non-canonical Staphylococcus aureus pathogenicity island repression. | | Descriptor: | Sri, Stl | | Authors: | Miguel-Romero, L, Alqasmi, M, Bacarizo, J, Tan, J.A, Cogdell, R.J, Chen, J, Byron, O, Christie, G.E, Marina, A, Penades, J.R. | | Deposit date: | 2021-07-10 | | Release date: | 2022-07-27 | | Last modified: | 2022-11-16 | | Method: | X-RAY DIFFRACTION (2.901 Å) | | Cite: | Non-canonical Staphylococcus aureus pathogenicity island repression.
Nucleic Acids Res., 50, 2022
|
|
5CCT
 
 | | Staphylococcus bacteriophage 80alpha dUTPase G164S mutant with dUpNHpp. | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, DUTPase, MAGNESIUM ION | | Authors: | Maiques, E, Quiles-Puchalt, N, Donderis, J, Ciges, J.R, Alite, C, Bowring, J, Penades, J.R, Marina, A. | | Deposit date: | 2015-07-02 | | Release date: | 2016-05-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Another look at the mechanism involving trimeric dUTPases in Staphylococcus aureus pathogenicity island induction involves novel players in the party.
Nucleic Acids Res., 44, 2016
|
|
5CCO
 
 | | Staphylococcus bacteriophage 80alpha dUTPase with dUMP. | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, DUTPase, MAGNESIUM ION | | Authors: | Maiques, E, Quiles-Puchalt, N, Donderis, J, Ciges, J.R, Alite, C, Bowring, J, Penades, J.R, Marina, A. | | Deposit date: | 2015-07-02 | | Release date: | 2016-05-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Another look at the mechanism involving trimeric dUTPases in Staphylococcus aureus pathogenicity island induction involves novel players in the party.
Nucleic Acids Res., 44, 2016
|
|
7QDG
 
 | | SARS-CoV-2 S protein S:A222V + S:D614G mutant 1-up | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ginex, T, Marco-Marin, C, Wieczor, M, Mata, C.P, Krieger, J, Lopez-Redondo, M.L, Frances-Gomez, C, Ruiz-Rodriguez, P, Melero, R, Sanchez-Sorzano, C.O, Martinez, M, Gougeard, N, Forcada-Nadal, A, Zamora-Caballero, S, Gozalbo-Rovira, R, Sanz-Frasquet, C, Bravo, J, Rubio, V, Marina, A, Geller, R, Comas, I, Gil, C, Coscolla, M, Orozco, M, LLacer, J.L, Carazo, J.M. | | Deposit date: | 2021-11-27 | | Release date: | 2022-05-25 | | Last modified: | 2022-08-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The structural role of SARS-CoV-2 genetic background in the emergence and success of spike mutations: The case of the spike A222V mutation.
Plos Pathog., 18, 2022
|
|
7QDH
 
 | | SARS-CoV-2 S protein S:D614G mutant 1-up | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin | | Authors: | Ginex, T, Marco-Marin, C, Wieczor, M, Mata, C.P, Krieger, J, Lopez-Redondo, M.L, Frances-Gomez, C, Ruiz-Rodriguez, P, Melero, R, Sanchez-Sorzano, C.O, Martinez, M, Gougeard, N, Forcada-Nadal, A, Zamora-Caballero, S, Gozalbo-Rovira, R, Sanz-Frasquet, C, Bravo, J, Rubio, V, Marina, A, Geller, R, Comas, I, Gil, C, Coscolla, M, Orozco, M, LLacer, J.L, Carazo, J.M. | | Deposit date: | 2021-11-27 | | Release date: | 2022-05-25 | | Last modified: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | The structural role of SARS-CoV-2 genetic background in the emergence and success of spike mutations: The case of the spike A222V mutation.
Plos Pathog., 18, 2022
|
|
5A19
 
 | | The structure of MAT2A in complex with PPNP. | | Descriptor: | (DIPHOSPHONO)AMINOPHOSPHONIC ACID, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Murray, B, Antonyuk, S.V, Marina, A, Lu, S.C, Mato, J.M, Hasnain, S.S, Rojas, A.L. | | Deposit date: | 2015-04-28 | | Release date: | 2016-02-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Crystallography Captures Catalytic Steps in Human Methionine Adenosyltransferase Enzymes.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
3DGF
 
 | |
