4ZJA
 
 | |
4ZJD
 
 | |
4ZJ9
 
 | |
7RX0
 
 | | Complex of AMPPNP-Kif7 and Gli2 Zinc-Finger domain bound to microtubules | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Kinesin-like protein KIF7, MAGNESIUM ION, ... | | Authors: | Mani, N, Wilson-Kubalek, E.M, Haque, F, Freniere, C, Milligan, R.A, Subramanian, R. | | Deposit date: | 2021-08-20 | | Release date: | 2022-06-15 | | Last modified: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (3.89 Å) | | Cite: | Cytoskeletal regulation of a transcription factor by DNA mimicry via coiled-coil interactions.
Nat.Cell Biol., 24, 2022
|
|
6MLR
 
 | | Cryo-EM structure of microtubule-bound Kif7 in the AMPPNP state | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, Kinesin-like protein KIF7, ... | | Authors: | Mani, N, Jiang, S, Wilson-Kubalek, E.M, Ku, P, Milligan, R.A, Subramanian, R. | | Deposit date: | 2018-09-27 | | Release date: | 2019-05-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Interplay between the Kinesin and Tubulin Mechanochemical Cycles Underlies Microtubule Tip Tracking by the Non-motile Ciliary Kinesin Kif7.
Dev.Cell, 49, 2019
|
|
6MLQ
 
 | | Cryo-EM structure of microtubule-bound Kif7 in the ADP state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Mani, N, Jiang, S, Wilson-Kubalek, E.M, Ku, P, Milligan, R.A, Subramanian, R. | | Deposit date: | 2018-09-27 | | Release date: | 2019-05-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Interplay between the Kinesin and Tubulin Mechanochemical Cycles Underlies Microtubule Tip Tracking by the Non-motile Ciliary Kinesin Kif7.
Dev.Cell, 49, 2019
|
|
5WQU
 
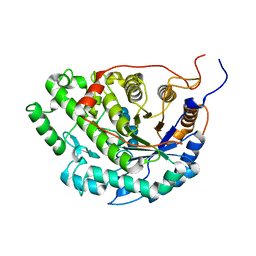 | | Crystal structure of Sweet Potato Beta-Amylase complexed with Maltotetraose | | Descriptor: | Beta-amylase, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Vajravijayan, S, Sergei, P, Nandhagopal, N, Gunasekaran, K. | | Deposit date: | 2016-11-28 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural insights on starch hydrolysis by plant beta-amylase and its evolutionary relationship with bacterial enzymes
Int. J. Biol. Macromol., 113, 2018
|
|
5WQS
 
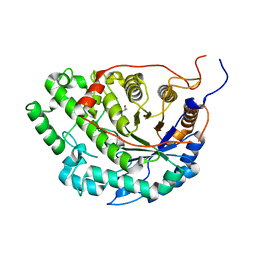 | | Crystal structure of Apo Beta-Amylase from Sweet potato | | Descriptor: | Beta-amylase, ISOPROPYL ALCOHOL | | Authors: | Vajravijayan, S, Sergei, P, Nandhagopal, N, Gunasekaran, K. | | Deposit date: | 2016-11-28 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights on starch hydrolysis by plant beta-amylase and its evolutionary relationship with bacterial enzymes
Int. J. Biol. Macromol., 113, 2018
|
|
8GBU
 
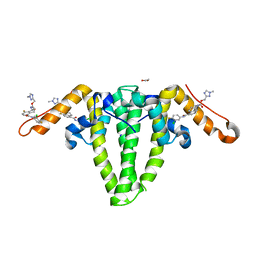 | | Hepatitis B capsid Y132A mutant with compound AB-506 | | Descriptor: | (1-methyl-1H-1,2,4-triazol-3-yl)methyl {(1S)-4-[(3-chloro-4-fluorophenyl)carbamoyl]-7-fluoro-2,3-dihydro-1H-inden-1-yl}carbamate, 1,2-ETHANEDIOL, Capsid protein | | Authors: | Horanyi, P.S, Mayclin, S.J. | | Deposit date: | 2023-02-28 | | Release date: | 2023-09-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Design, synthesis, and structure-activity relationship of a bicyclic HBV capsid assembly modulator chemotype leading to the identification of clinical candidate AB-506.
Bioorg.Med.Chem.Lett., 94, 2023
|
|
7S76
 
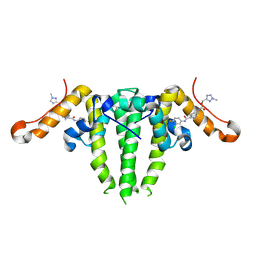 | | HBV CAPSID Y132A WITH COMPOUND 10b AT 2.5A RESOLUTION | | Descriptor: | (1-methyl-1H-1,2,4-triazol-3-yl)methyl {(4S)-1-[(3-chloro-4-fluorophenyl)carbamoyl]-2-methyl-2,4,5,6-tetrahydrocyclopenta[c]pyrrol-4-yl}carbamate, Capsid protein | | Authors: | Olland, A.M, Suto, R.K. | | Deposit date: | 2021-09-15 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The identification of highly efficacious functionalised tetrahydrocyclopenta[ c ]pyrroles as inhibitors of HBV viral replication through modulation of HBV capsid assembly.
Rsc Med Chem, 13, 2022
|
|
3G7B
 
 | |
3G7E
 
 | |
3G75
 
 | |
