4R47
 
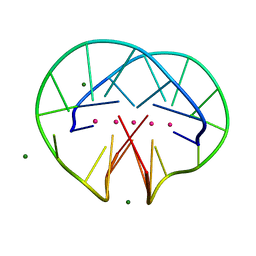 | | Racemic crystal structure of a bimolecular DNA G-quadruplex (P21/n) | | Descriptor: | 5'-D(*GP*GP*GP*GP*TP*TP*TP*TP*GP*GP*GP*G)-3', MAGNESIUM ION, POTASSIUM ION | | Authors: | Mandal, P.K, Collie, G.W, Kauffmann, B, Huc, I. | | Deposit date: | 2014-08-19 | | Release date: | 2014-11-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Racemic DNA crystallography.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4R44
 
 | | Racemic crystal structure of a tetramolecular DNA G-quadruplex | | Descriptor: | 5'-D(*TP*GP*GP*GP*GP*T)-3', POTASSIUM ION, SODIUM ION | | Authors: | Mandal, P.K, Collie, G.W, Kauffmann, B, Huc, I. | | Deposit date: | 2014-08-19 | | Release date: | 2014-11-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.695 Å) | | Cite: | Racemic DNA crystallography.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
6GN2
 
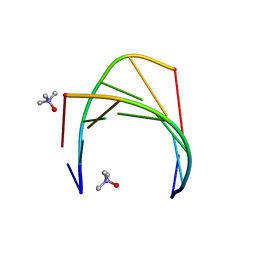 | |
6GN3
 
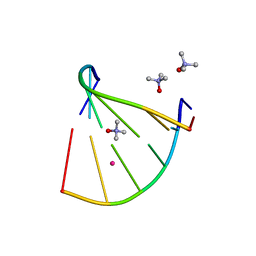 | | Racemic crystal structure of A-DNA duplex formed from d(CCCGGG) in space group P21/n | | Descriptor: | CHLORIDE ION, COBALT (II) ION, DNA (5'-D(*CP*CP*CP*GP*GP*G)-3'), ... | | Authors: | Mandal, P.K, Collie, G.W, Kauffmann, B, Huc, I. | | Deposit date: | 2018-05-29 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Racemic crystal structures of A-DNA duplexes
To Be Published
|
|
5J0E
 
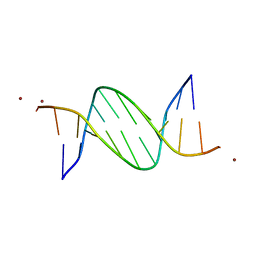 | | Crystal structures of Pribnow box consensus promoter sequence (P32) | | Descriptor: | Complementary sequence, Pribnow box consensus promoter sequence, ZINC ION | | Authors: | Mandal, P.K, Collie, G.W, Kauffmann, B, Huc, I. | | Deposit date: | 2016-03-28 | | Release date: | 2016-05-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Structure elucidation of the Pribnow box consensus promoter sequence by racemic DNA crystallography.
Nucleic Acids Res., 44, 2016
|
|
5EYQ
 
 | | Racemic crystal structures of Pribnow box consensus promoter sequence (Pnna) | | Descriptor: | Complementary strand, Pribnow box template strand, SODIUM ION | | Authors: | Mandal, P.K, Collie, G.W, Kauffmann, B, Srivastava, S.C, Huc, I. | | Deposit date: | 2015-11-25 | | Release date: | 2016-05-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure elucidation of the Pribnow box consensus promoter sequence by racemic DNA crystallography.
Nucleic Acids Res., 44, 2016
|
|
5EZF
 
 | | Racemic crystal structures of Pribnow box consensus promoter sequence (Pbca) | | Descriptor: | CALCIUM ION, Complementary strand, Pribnow box template strand | | Authors: | Mandal, P.K, Collie, G.W, Kauffmann, B, Srivastava, S.C, Huc, I. | | Deposit date: | 2015-11-26 | | Release date: | 2016-05-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure elucidation of the Pribnow box consensus promoter sequence by racemic DNA crystallography.
Nucleic Acids Res., 44, 2016
|
|
5F26
 
 | | Crystal structures of Pribnow box consensus promoter sequence (P63) | | Descriptor: | Complementary strand, Pribnow box consensus sequence strand | | Authors: | Mandal, P.K, Collie, G.W, Kauffmann, B, Srivastava, S.C, Huc, I. | | Deposit date: | 2015-12-01 | | Release date: | 2016-05-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure elucidation of the Pribnow box consensus promoter sequence by racemic DNA crystallography.
Nucleic Acids Res., 44, 2016
|
|
5EWB
 
 | | Racemic crystal structures of Pribnow box consensus promoter sequence (P21/c) | | Descriptor: | PRIBNOW BOX CONSENSUS SEQUENCE- NON-TEMPLATE STRAND, PRIBNOW BOX CONSENSUS SEQUENCE- TEMPLATE STRAND | | Authors: | Mandal, P.K, Collie, G.W, Kauffmann, B, Srivastava, S.C, Huc, I. | | Deposit date: | 2015-11-20 | | Release date: | 2016-05-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.694 Å) | | Cite: | Structure elucidation of the Pribnow box consensus promoter sequence by racemic DNA crystallography.
Nucleic Acids Res., 44, 2016
|
|
3T8P
 
 | |
3ULM
 
 | |
3ULO
 
 | |
3UM4
 
 | |
3ULN
 
 | |
3V9D
 
 | |
4DWY
 
 | | Interactions of Mn2+ with a non-self-complementary Z-type DNA duplex | | Descriptor: | DNA (5'-D(*CP*AP*CP*GP*CP*G)-3'), DNA (5'-D(*CP*GP*CP*GP*TP*G)-3'), MANGANESE (II) ION | | Authors: | Mandal, P.K, Venkadesh, S, Gautham, N. | | Deposit date: | 2012-02-27 | | Release date: | 2012-11-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Interactions of Mn(2+) with a non-self-complementary Z-type DNA duplex
Acta Crystallogr.,Sect.F, 68, 2012
|
|
4DY8
 
 | | Interactions of Mn2+ with a non-self-complementary Z-type DNA duplex | | Descriptor: | DNA (5'-D(*CP*AP*CP*GP*CP*G)-3'), DNA (5'-D(*CP*GP*CP*GP*TP*G)-3'), DNA (5'-D(P*TP*G)-3'), ... | | Authors: | Mandal, P.K, Venkadesh, S, Gautham, N. | | Deposit date: | 2012-02-28 | | Release date: | 2012-11-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Interactions of Mn(2+) with a non-self-complementary Z-type DNA duplex
Acta Crystallogr.,Sect.F, 68, 2012
|
|
4E2R
 
 | |
4E60
 
 | |
4E4O
 
 | |
5HIX
 
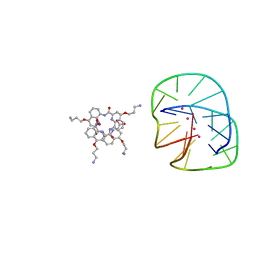 | | Cocrystal structure of an anti-parallel DNA G-quadruplex and a tetra-Quinoline Foldamer | | Descriptor: | 4-(3-aminopropoxy)-8-({[4-(3-aminopropoxy)-8-({[4-(3-aminopropoxy)-8-({[4-(3-aminopropoxy)-8-nitroquinolin-2-yl]carbonyl}amino)quinolin-2-yl]carbonyl}amino)quinolin-2-yl]carbonyl}amino)quinoline-2-carboxylic acid, Dimeric G-quadruplex, POTASSIUM ION | | Authors: | Mandal, P.K, Baptiste, B, Langlois d'Estaintot, B, Kauffmann, B, Huc, I. | | Deposit date: | 2016-01-12 | | Release date: | 2016-11-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Multivalent Interactions between an Aromatic Helical Foldamer and a DNA G-Quadruplex in the Solid State.
Chembiochem, 17, 2016
|
|
5ET9
 
 | | Racemic crystal structures of Pribnow box consensus promoter sequence (P21/n) | | Descriptor: | BARIUM ION, Pribnow box consensus sequence- template strand, Pribnow box non-template strand | | Authors: | Mandal, P.K, Collie, G.W, Kauffmann, B, Srivastava, S.C, Huc, I. | | Deposit date: | 2015-11-17 | | Release date: | 2016-05-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structure elucidation of the Pribnow box consensus promoter sequence by racemic DNA crystallography.
Nucleic Acids Res., 44, 2016
|
|
4R45
 
 | | Racemic crystal structure of a bimolecular DNA G-quadruplex (P-1) | | Descriptor: | 5'-D(*GP*GP*GP*GP*TP*TP*TP*TP*GP*GP*GP*G)-3', POTASSIUM ION | | Authors: | Mandal, P.K, Collie, G.W, Kauffmann, B, Huc, I. | | Deposit date: | 2014-08-19 | | Release date: | 2014-11-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Racemic DNA crystallography.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4R4A
 
 | | Racemic crystal structure of a cobalt-bound B-DNA duplex | | Descriptor: | 5'-D(*CP*CP*GP*GP*TP*AP*CP*CP*GP*G)-3', COBALT (II) ION, SPERMINE | | Authors: | Mandal, P.K, Collie, G.W, Kauffmann, B, Huc, I. | | Deposit date: | 2014-08-19 | | Release date: | 2014-11-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Racemic DNA crystallography.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4R49
 
 | | Racemic crystal structure of a calcium-bound B-DNA duplex | | Descriptor: | 5'-D(*CP*CP*GP*GP*TP*AP*CP*CP*GP*G)-3', CALCIUM ION, SODIUM ION | | Authors: | Mandal, P.K, Collie, G.W, Kauffmann, B, Huc, I. | | Deposit date: | 2014-08-19 | | Release date: | 2014-11-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Racemic DNA crystallography.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
