2A5S
 
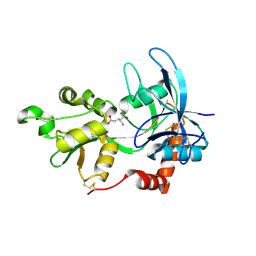 | | Crystal Structure Of The NR2A Ligand Binding Core In Complex With Glutamate | | Descriptor: | GLUTAMIC ACID, N-methyl-D-aspartate receptor NMDAR2A subunit | | Authors: | Furukawa, H, Singh, S.K, Mancusso, R, Gouaux, E. | | Deposit date: | 2005-06-30 | | Release date: | 2005-11-15 | | Last modified: | 2017-07-26 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Subunit arrangement and function in NMDA receptors
Nature, 438, 2005
|
|
2A5T
 
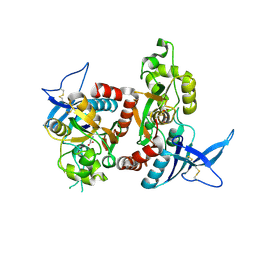 | | Crystal Structure Of The NR1/NR2A ligand-binding cores complex | | Descriptor: | GLUTAMIC ACID, GLYCINE, N-methyl-D-aspartate receptor NMDAR1-4a subunit, ... | | Authors: | Furukawa, H, Singh, S.K, Mancusso, R, Gouaux, E. | | Deposit date: | 2005-06-30 | | Release date: | 2005-11-15 | | Last modified: | 2017-07-26 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Subunit arrangement and function in NMDA receptors
Nature, 438, 2005
|
|
4F35
 
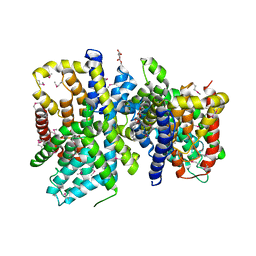 | | Crystal Structure of a bacterial dicarboxylate/sodium symporter | | Descriptor: | CITRIC ACID, SODIUM ION, Transporter, ... | | Authors: | Mancusso, R.L, Gregorio, G.G, Liu, Q, Wang, D.N. | | Deposit date: | 2012-05-08 | | Release date: | 2012-10-24 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.196 Å) | | Cite: | Structure and mechanism of a bacterial sodium-dependent dicarboxylate transporter.
Nature, 491, 2012
|
|
