6DK8
 
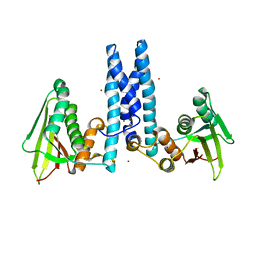 | | RetS kinase region without cobalt | | Descriptor: | NICKEL (II) ION, RetS (Regulator of Exopolysaccharide and Type III Secretion) | | Authors: | Mancl, J.M, Schubot, F.D. | | Deposit date: | 2018-05-29 | | Release date: | 2019-03-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Helix Cracking Regulates the Critical Interaction between RetS and GacS in Pseudomonas aeruginosa.
Structure, 27, 2019
|
|
6DK7
 
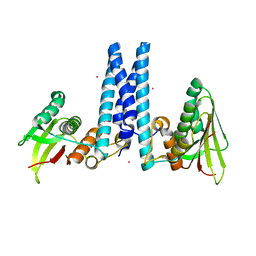 | | RetS histidine kinase region with cobalt | | Descriptor: | COBALT (II) ION, RetS (Regulator of Exopolysaccharide and Type III Secretion) | | Authors: | Mancl, J.M, Schubot, F.D. | | Deposit date: | 2018-05-29 | | Release date: | 2019-03-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Helix Cracking Regulates the Critical Interaction between RetS and GacS in Pseudomonas aeruginosa.
Structure, 27, 2019
|
|
7RZI
 
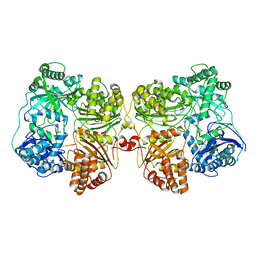 | | Insulin Degrading Enzyme pC/pC | | Descriptor: | Cysteine-free Insulin-degrading enzyme, Insulin A chain, Insulin B chain | | Authors: | Mancl, J.M, Liang, W.G, Tang, W.J. | | Deposit date: | 2021-08-27 | | Release date: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Ensemble cryoEM reveals a substrate-induced shift in the conformational dynamics of human insulin degrading enzyme
To be published
|
|
7RZE
 
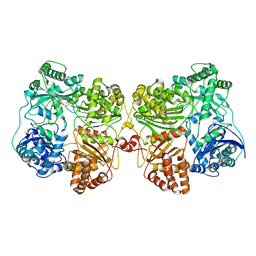 | | Insulin Degrading Enzyme pO/pC | | Descriptor: | Cysteine-free Insulin-degrading enzyme, Insulin A chain, Insulin B chain | | Authors: | Mancl, J.M, Liang, W.G, Tang, W.J. | | Deposit date: | 2021-08-27 | | Release date: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Ensemble cryoEM reveals a substrate-induced shift in the conformational dynamics of human insulin degrading enzyme
To be published
|
|
7RZH
 
 | | Insulin Degrading Enzyme O/O | | Descriptor: | Cysteine-free Insulin-degrading enzyme | | Authors: | Mancl, J.M, Liang, W.G, Tang, W.J. | | Deposit date: | 2021-08-27 | | Release date: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Ensemble cryoEM reveals a substrate-induced shift in the conformational dynamics of human insulin degrading enzyme
To be published
|
|
7RZG
 
 | | Insulin Degrading Enzyme O/pO | | Descriptor: | Cysteine-free Insulin-degrading enzyme | | Authors: | Mancl, J.M, Liang, W.G, Tang, W.J. | | Deposit date: | 2021-08-27 | | Release date: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Ensemble cryoEM reveals a substrate-induced shift in the conformational dynamics of human insulin degrading enzyme
To be published
|
|
7RZF
 
 | | Insulin Degrading Enzyme O/pC | | Descriptor: | Cysteine-free Insulin-degrading enzyme, Insulin A chain, Insulin B chain | | Authors: | Mancl, J.M, Liang, W.G, Tang, W.J. | | Deposit date: | 2021-08-27 | | Release date: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Ensemble cryoEM reveals a substrate-induced shift in the conformational dynamics of human insulin degrading enzyme
To be published
|
|
7K1V
 
 | |
5IT5
 
 | | Thermus thermophilus PilB core ATPase region | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP binding motif-containing protein PilF, MAGNESIUM ION, ... | | Authors: | Mancl, J, Robinson, H, Black, W, Yang, Z, Schubot, F. | | Deposit date: | 2016-03-16 | | Release date: | 2016-10-19 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.648 Å) | | Cite: | Crystal Structure of a Type IV Pilus Assembly ATPase: Insights into the Molecular Mechanism of PilB from Thermus thermophilus.
Structure, 24, 2016
|
|
6XLY
 
 | |
6D8V
 
 | | Methyl-accepting Chemotaxis protein X | | Descriptor: | 1,1-DIMETHYL-PROLINIUM, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Probable chemoreceptor (Methyl-accepting chemotaxis) transmembrane protein | | Authors: | Shrestha, M, Schubot, F.D. | | Deposit date: | 2018-04-27 | | Release date: | 2019-04-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the sensory domain of McpX fromSinorhizobium meliloti, the first known bacterial chemotactic sensor for quaternary ammonium compounds.
Biochem. J., 475, 2018
|
|
6XOS
 
 | |
6XOU
 
 | |
6XOV
 
 | |
6XOT
 
 | |
