7REQ
 
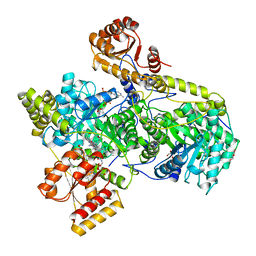 | |
6REQ
 
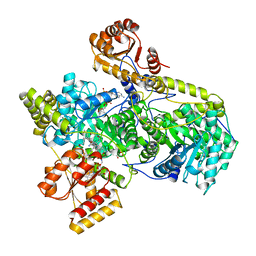 | |
3REQ
 
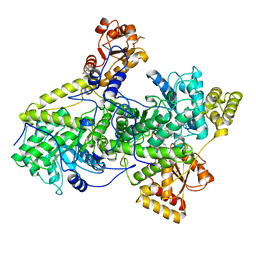 | |
4REQ
 
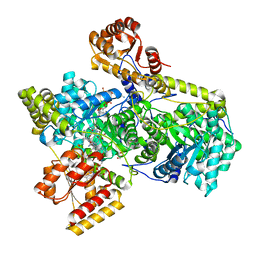 | | Methylmalonyl-COA Mutase substrate complex | | Descriptor: | 5'-DEOXYADENOSINE, COBALAMIN, GLYCEROL, ... | | Authors: | Evans, P.R, Mancia, F. | | Deposit date: | 1998-06-17 | | Release date: | 1999-01-13 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Conformational changes on substrate binding to methylmalonyl CoA mutase and new insights into the free radical mechanism.
Structure, 6, 1998
|
|
1REQ
 
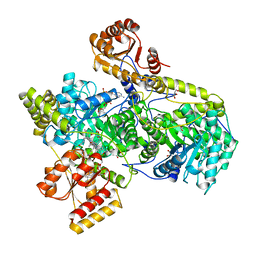 | | METHYLMALONYL-COA MUTASE | | Descriptor: | COBALAMIN, DESULFO-COENZYME A, GLYCEROL, ... | | Authors: | Evans, P.R, Mancia, F. | | Deposit date: | 1996-01-19 | | Release date: | 1997-01-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | How coenzyme B12 radicals are generated: the crystal structure of methylmalonyl-coenzyme A mutase at 2 A resolution.
Structure, 4, 1996
|
|
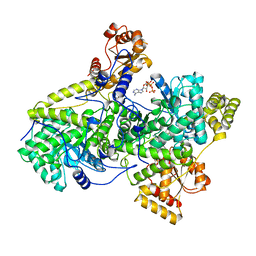
2REQ
 
 | |
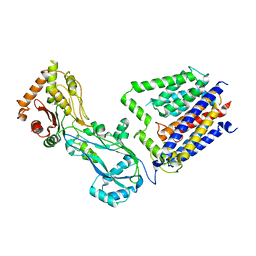
8TJ3
 
 | |
7TPJ
 
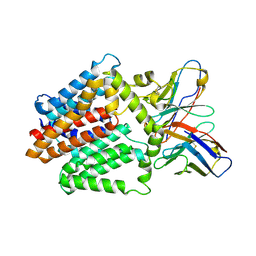 | | Single-Particle Cryo-EM Structure of the WaaL O-antigen ligase in its apo state | | Descriptor: | Fab Heavy (H) Chain, Fab Light (L) Chain, Putative cell surface polysaccharide polymerase/ligase | | Authors: | Ashraf, K.U, Nygaard, R, Vickery, O.N, Erramilli, S.K, Herrera, C.M, McConville, T.H, Petrou, V.I, Giacometti, S.I, Dufrisne, M.B, Nosol, K, Zinkle, A.P, Graham, C.L.B, Loukeris, M, Kloss, B, Skorupinska-Tudek, K, Swiezewska, E, Roper, D, Clarke, O.B, Uhlemann, A.C, Kossiakoff, A.A, Trent, M.S, Stansfeld, P.J, Mancia, F. | | Deposit date: | 2022-01-25 | | Release date: | 2022-04-06 | | Last modified: | 2022-04-27 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Structural basis of lipopolysaccharide maturation by the O-antigen ligase.
Nature, 604, 2022
|
|
7TPG
 
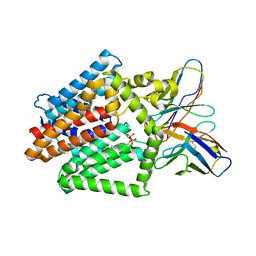 | | Single-Particle Cryo-EM Structure of the WaaL O-antigen ligase in its ligand bound state | | Descriptor: | Fab Heavy (H) Chain, Fab Light (L) Chain, GERANYL DIPHOSPHATE, ... | | Authors: | Ashraf, K.U, Nygaard, R, Vickery, O.N, Erramilli, S.K, Herrera, C.M, McConville, T.H, Petrou, V.I, Giacometti, S.I, Dufrisne, M.B, Nosol, K, Zinkle, A.P, Graham, C.L.B, Loukeris, M, Kloss, B, Skorupinska-Tudek, K, Swiezewska, E, Roper, D, Clarke, O.B, Uhlemann, A.C, Kossiakoff, A.A, Trent, M.S, Stansfeld, P.J, Mancia, F. | | Deposit date: | 2022-01-25 | | Release date: | 2022-04-06 | | Last modified: | 2022-04-27 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Structural basis of lipopolysaccharide maturation by the O-antigen ligase.
Nature, 604, 2022
|
|
5K8Q
 
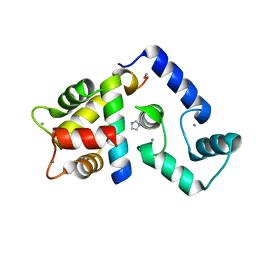 | | Crystal Structure of Calcium-loaded Calmodulin in complex with STRA6 CaMBP2-site peptide. | | Descriptor: | CALCIUM ION, Calmodulin, IMIDAZOLE, ... | | Authors: | Stowe, S.D, Clarke, O.B, Cavalier, M.C, Godoy-Ruiz, R, Mancia, F, Weber, D.J. | | Deposit date: | 2016-05-30 | | Release date: | 2016-08-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.739 Å) | | Cite: | Structure of the STRA6 receptor for retinol uptake.
Science, 353, 2016
|
|
1HQP
 
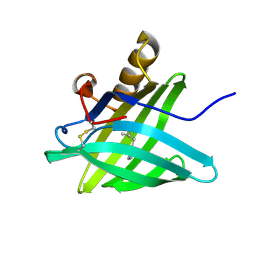 | | CRYSTAL STRUCTURE OF A TRUNCATED FORM OF PORCINE ODORANT-BINDING PROTEIN | | Descriptor: | 2-ISOBUTYL-3-METHOXYPYRAZINE, ODORANT-BINDING PROTEIN | | Authors: | Perduca, M, Mancia, F, Del Giorgio, R, Monaco, H.L. | | Deposit date: | 2000-12-19 | | Release date: | 2001-01-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a truncated form of porcine odorant-binding protein.
Proteins, 42, 2001
|
|
5SY1
 
 | | Structure of the STRA6 receptor for retinol uptake in complex with calmodulin | | Descriptor: | CALCIUM ION, CHOLESTEROL, Calmodulin, ... | | Authors: | Clarke, O.B, Chen, Y, Mancia, F. | | Deposit date: | 2016-08-10 | | Release date: | 2016-08-24 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure of the STRA6 receptor for retinol uptake.
Science, 353, 2016
|
|
3TRK
 
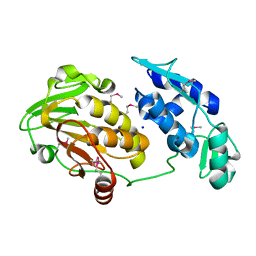 | | Structure of the Chikungunya virus nsP2 protease | | Descriptor: | Nonstructural polyprotein, SODIUM ION | | Authors: | Cheung, J, Franklin, M, Mancia, F, Rudolph, M, Cassidy, M, Gary, E, Burshteyn, F, Love, J. | | Deposit date: | 2011-09-09 | | Release date: | 2011-09-28 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.397 Å) | | Cite: | Structure of the Chikungunya virus nsP2 protease
To be Published
|
|
7NP4
 
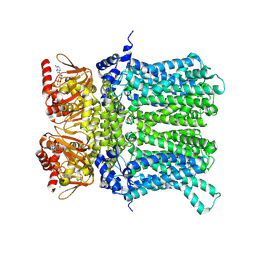 | | cAMP-bound rabbit HCN4 stabilized in LMNG-CHS detergent mixture | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4,Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 | | Authors: | Giese, H, Chaves-Sanjuan, A, Saponaro, A, Clarke, O, Bolognesi, M, Mancia, F, Hendrickson, W.A, Thiel, G, Santoro, B, Moroni, A. | | Deposit date: | 2021-02-26 | | Release date: | 2021-08-11 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Gating movements and ion permeation in HCN4 pacemaker channels.
Mol.Cell, 81, 2021
|
|
7NP3
 
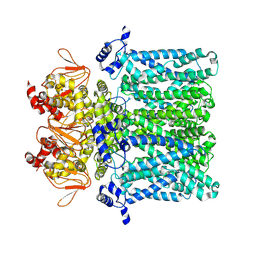 | | cAMP-free rabbit HCN4 stabilized in LMNG-CHS detergent mixture | | Descriptor: | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4,Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 | | Authors: | Giese, H.M, Chaves-Sanjuan, A, Saponaro, A, Clarke, O, Bolognesi, M, Mancia, F, Hendrickson, W.A, Thiel, G, Santoro, B, Moroni, A. | | Deposit date: | 2021-02-26 | | Release date: | 2021-08-11 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Gating movements and ion permeation in HCN4 pacemaker channels.
Mol.Cell, 81, 2021
|
|
7KC4
 
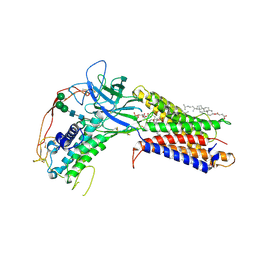 | | Human WLS in complex with WNT8A | | Descriptor: | 1-CIS-9-OCTADECANOYL-2-CIS-9-HEXADECANOYL PHOSPHATIDYL GLYCEROL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Nygaard, R, Jia, Y, Kim, J, Ross, D, Parisi, G, Clarke, O.B, Virshup, D.M, Mancia, F. | | Deposit date: | 2020-10-05 | | Release date: | 2021-01-06 | | Last modified: | 2021-01-20 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Structural Basis of WLS/Evi-Mediated Wnt Transport and Secretion.
Cell, 184, 2021
|
|
4O6M
 
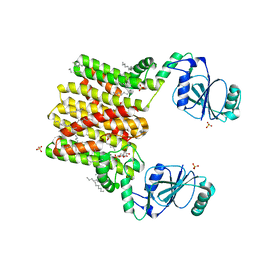 | | Structure of AF2299, a CDP-alcohol phosphotransferase (CMP-bound) | | Descriptor: | AF2299, a CDP-alcohol phosphotransferase, CALCIUM ION, ... | | Authors: | Clarke, O.B, Sciara, G, Tomasek, D, Banerjee, S, Rajashankar, K.R, Shapiro, L, Mancia, F, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2013-12-22 | | Release date: | 2014-05-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Structural basis for catalysis in a CDP-alcohol phosphotransferase.
Nat Commun, 5, 2014
|
|
4O6N
 
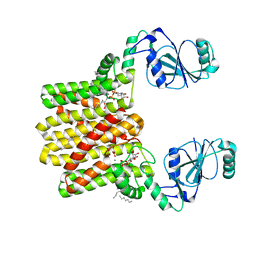 | | Structure of AF2299, a CDP-alcohol phosphotransferase (CDP-bound) | | Descriptor: | AF2299, a CDP-alcohol phosphotransferase, CALCIUM ION, ... | | Authors: | Clarke, O.B, Sciara, G, Tomasek, D, Banerjee, S, Rajashankar, K.R, Shapiro, L, Mancia, F, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2013-12-22 | | Release date: | 2014-05-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for catalysis in a CDP-alcohol phosphotransferase.
Nat Commun, 5, 2014
|
|
4Q7C
 
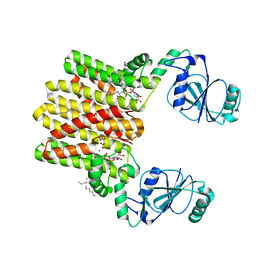 | | Structure of AF2299, a CDP-alcohol phosphotransferase | | Descriptor: | AF2299, a CDP-alcohol phosphotransferase, CALCIUM ION, ... | | Authors: | Clarke, O.B, Sciara, G, Tomasek, D, Banerjee, S, Rajashankar, K.R, Shapiro, L, Mancia, F, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2014-04-24 | | Release date: | 2014-05-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.102 Å) | | Cite: | Structural basis for catalysis in a CDP-alcohol phosphotransferase.
Nat Commun, 5, 2014
|
|
3TX3
 
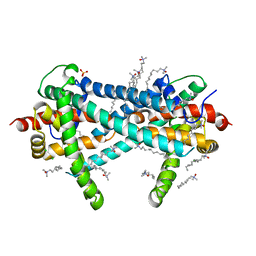 | | CysZ, a putative sulfate permease | | Descriptor: | CHLORIDE ION, LAURYL DIMETHYLAMINE-N-OXIDE, SULFATE ION, ... | | Authors: | Assur, Z, Liu, Q, Hendrickson, W.A, Mancia, F, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2011-09-22 | | Release date: | 2011-11-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | CysZ, a putative sulfate permease
To be Published
|
|
7MJS
 
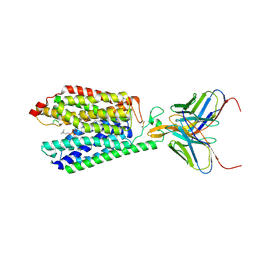 | | Single-Particle Cryo-EM Structure of Major Facilitator Superfamily Domain containing 2A in complex with LPC-18:3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2AG3 Fab heavy chain, 2AG3 Fab light chain, ... | | Authors: | Cater, R.J, Chua, G.L, Erramilli, S.K, Keener, J.E, Choy, B.C, Tokarz, P, Chin, C.F, Quek, D.Q.Y, Kloss, B, Pepe, J.G, Parisi, G, Wong, B.H, Clarke, O.B, Marty, M.T, Kossiakoff, A.A, Khelashvili, G, Silver, D.L, Mancia, F. | | Deposit date: | 2021-04-20 | | Release date: | 2021-06-16 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Structural basis of omega-3 fatty acid transport across the blood-brain barrier.
Nature, 595, 2021
|
|
6UKJ
 
 | | Single-Particle Cryo-EM Structure of Plasmodium falciparum Chloroquine Resistance Transporter (PfCRT) 7G8 Isoform | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Chloroquine resistance transporter, Fab Heavy Chain, ... | | Authors: | Kim, J, Tan, Y.Z, Wicht, K.J, Erramilli, S.K, Dhingra, S.K, Okombo, J, Vendome, J, Hagenah, L.M, Giacometti, S.I, Warren, A.L, Nosol, K, Roepe, P.D, Potter, C.S, Carragher, B, Kossiakoff, A.A, Quick, M, Fidock, D.A, Mancia, F. | | Deposit date: | 2019-10-05 | | Release date: | 2019-12-04 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure and drug resistance of the Plasmodium falciparum transporter PfCRT.
Nature, 576, 2019
|
|
6W98
 
 | | Single-Particle Cryo-EM Structure of Arabinofuranosyltransferase AftD from Mycobacteria | | Descriptor: | 4'-PHOSPHOPANTETHEINE, Acyl carrier protein, CALCIUM ION, ... | | Authors: | Tan, Y.Z, Zhang, L, Rodrigues, J, Zheng, R.B, Giacometti, S.I, Rosario, A.L, Kloss, B, Dandey, V.P, Wei, H, Brunton, R, Raczkowski, A.M, Athayde, D, Catalao, M.J, Pimentel, M, Clarke, O.B, Lowary, T.L, Archer, M, Niederweis, M, Potter, C.S, Carragher, B, Mancia, F. | | Deposit date: | 2020-03-22 | | Release date: | 2020-05-13 | | Last modified: | 2020-06-03 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM Structures and Regulation of Arabinofuranosyltransferase AftD from Mycobacteria.
Mol.Cell, 78, 2020
|
|
6WBX
 
 | | Single-Particle Cryo-EM Structure of Arabinofuranosyltransferase AftD from Mycobacteria, Mutant R1389S Class 1 | | Descriptor: | CALCIUM ION, DUF3367 domain-containing protein | | Authors: | Tan, Y.Z, Zhang, L, Rodrigues, J, Zheng, R.B, Giacometti, S.I, Rosario, A.L, Kloss, B, Dandey, V.P, Wei, H, Brunton, R, Raczkowski, A.M, Athayde, D, Catalao, M.J, Pimentel, M, Clarke, O.B, Lowary, T.L, Archer, M, Niederweis, M, Potter, C.S, Carragher, B, Mancia, F. | | Deposit date: | 2020-03-27 | | Release date: | 2020-05-13 | | Last modified: | 2020-06-03 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM Structures and Regulation of Arabinofuranosyltransferase AftD from Mycobacteria.
Mol.Cell, 78, 2020
|
|
6WBY
 
 | | Single-Particle Cryo-EM Structure of Arabinofuranosyltransferase AftD from Mycobacteria, Mutant R1389S Class 2 | | Descriptor: | CALCIUM ION, DUF3367 domain-containing protein | | Authors: | Tan, Y.Z, Zhang, L, Rodrigues, J, Zheng, R.B, Giacometti, S.I, Rosario, A.L, Kloss, B, Dandey, V.P, Wei, H, Brunton, R, Raczkowski, A.M, Athayde, D, Catalao, M.J, Pimentel, M, Clarke, O.B, Lowary, T.L, Archer, M, Niederweis, M, Potter, C.S, Carragher, B, Mancia, F. | | Deposit date: | 2020-03-27 | | Release date: | 2020-05-13 | | Last modified: | 2020-06-03 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM Structures and Regulation of Arabinofuranosyltransferase AftD from Mycobacteria.
Mol.Cell, 78, 2020
|
|
