6CI9
 
 | |
5SUH
 
 | |
6CI8
 
 | |
6EF6
 
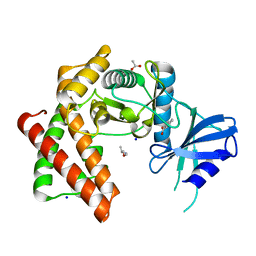 | | Structure of the microcompartment-associated aminopropanol kinase | | Descriptor: | (2R)-1-methoxypropan-2-amine, ACETATE ION, Aminoglycoside phosphotransferase, ... | | Authors: | Mallette, E, Kimber, M.S. | | Deposit date: | 2018-08-16 | | Release date: | 2018-10-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural and kinetic characterization of (S)-1-amino-2-propanol kinase from the aminoacetone utilization microcompartment ofMycobacterium smegmatis.
J.Biol.Chem., 293, 2018
|
|
5L38
 
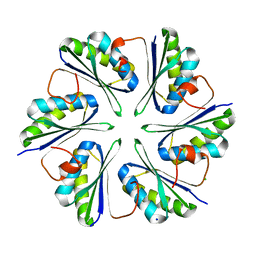 | |
5L37
 
 | |
5L39
 
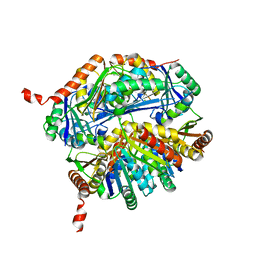 | |
5HNO
 
 | | The structure of the kdo-capped saccharide binding subunit of the O-12 specific ABC transporter, Wzt | | Descriptor: | ABC type transport system putative ATP binding protein, CHLORIDE ION | | Authors: | Mallette, E, Mann, E, Whitfield, C, Kimber, M.S. | | Deposit date: | 2016-01-18 | | Release date: | 2016-03-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Klebsiella pneumoniae O12 ATP-binding Cassette (ABC) Transporter Recognizes the Terminal Residue of Its O-antigen Polysaccharide Substrate.
J.Biol.Chem., 291, 2016
|
|
5HNP
 
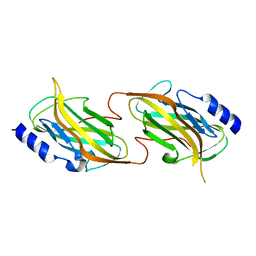 | | The structure of the kdo-capped saccharide binding subunit of the O-12 specific ABC transporter, Wzt | | Descriptor: | ABC transporter, CHLORIDE ION | | Authors: | Mallette, E, Mann, E, Whitfield, C, Kimber, M.S. | | Deposit date: | 2016-01-18 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Klebsiella pneumoniae O12 ATP-binding Cassette (ABC) Transporter Recognizes the Terminal Residue of Its O-antigen Polysaccharide Substrate.
J.Biol.Chem., 291, 2016
|
|
5FA0
 
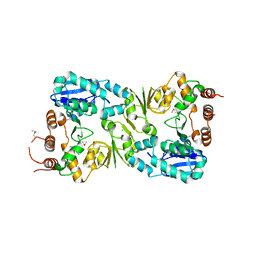 | | The structure of the beta-3-deoxy-D-manno-oct-2-ulosonic acid transferase domain from WbbB | | Descriptor: | CHLORIDE ION, Putative N-acetyl glucosaminyl transferase | | Authors: | Mallette, E, Ovchinnikova, O.G, Whitfield, C, Kimber, M.S. | | Deposit date: | 2015-12-10 | | Release date: | 2016-05-18 | | Last modified: | 2018-04-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Bacterial beta-Kdo glycosyltransferases represent a new glycosyltransferase family (GT99).
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
5FA1
 
 | | The structure of the beta-3-deoxy-D-manno-oct-2-ulosonic acid transferase domain of WbbB | | Descriptor: | CYTIDINE-5'-MONOPHOSPHATE, Putative N-acetyl glucosaminyl transferase | | Authors: | Mallette, E, Ovchinnikova, O.G, Whitfield, C, Kimber, M.S. | | Deposit date: | 2015-12-10 | | Release date: | 2016-05-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Bacterial beta-Kdo glycosyltransferases represent a new glycosyltransferase family (GT99).
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
6OK1
 
 | | Ltp2-ChsH2(DUF35) aldolase | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ChsH2(DUF35), ... | | Authors: | Kimber, M.S, Mallette, E, Aggett, R, Seah, S.Y.K. | | Deposit date: | 2019-04-12 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The steroid side-chain-cleaving aldolase Ltp2-ChsH2DUF35is a thiolase superfamily member with a radically repurposed active site.
J.Biol.Chem., 294, 2019
|
|
6MGC
 
 | | Escherichia coli KpsC, N-terminal domain | | Descriptor: | CHLORIDE ION, CYTIDINE-5'-MONOPHOSPHATE, Capsule polysaccharide export protein KpsC, ... | | Authors: | Doyle, L, Mallette, E, Kimber, M.S. | | Deposit date: | 2018-09-13 | | Release date: | 2019-03-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Biosynthesis of a conserved glycolipid anchor for Gram-negative bacterial capsules.
Nat.Chem.Biol., 15, 2019
|
|
6MGD
 
 | | Thermosulfurimonas dismutans KpsC, beta Kdo 2,7 transferase | | Descriptor: | Capsular polysaccharide export system protein KpsC | | Authors: | Doyle, L, Mallette, E, Kimber, M.S, Whitfield, C. | | Deposit date: | 2018-09-13 | | Release date: | 2019-03-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Biosynthesis of a conserved glycolipid anchor for Gram-negative bacterial capsules.
Nat.Chem.Biol., 15, 2019
|
|
6MGB
 
 | | Thermosulfurimonas dismutans KpsC, beta Kdo 2,4 transferase | | Descriptor: | CHLORIDE ION, CYTIDINE-5'-MONOPHOSPHATE, Capsular polysaccharide export system protein KpsC, ... | | Authors: | Doyle, L, Mallette, E, Kimber, M.S, Whitfield, C. | | Deposit date: | 2018-09-13 | | Release date: | 2019-03-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Biosynthesis of a conserved glycolipid anchor for Gram-negative bacterial capsules.
Nat.Chem.Biol., 15, 2019
|
|
6U4B
 
 | | WbbM bifunctional glycosytransferase apo structure | | Descriptor: | MAGNESIUM ION, WbbM protein | | Authors: | Kimber, M.S, Mallette, E, Kamski-Hennekam, E.R, Gitalis, R. | | Deposit date: | 2019-08-25 | | Release date: | 2020-01-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A bifunctional O-antigen polymerase structure reveals a new glycosyltransferase family.
Nat.Chem.Biol., 16, 2020
|
|
7L97
 
 | | Crystal structure of STAMBPL1 in complex with an engineered binder | | Descriptor: | 1,2-ETHANEDIOL, AMSH-like protease, SULFATE ION, ... | | Authors: | Guo, Y, Dong, A, Hou, F, Li, Y, Zhang, W, Arrowsmith, C.H, Edwards, A.M, Tong, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-01-02 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural and functional characterization of ubiquitin variant inhibitors for the JAMM-family deubiquitinases STAMBP and STAMBPL1.
J.Biol.Chem., 297, 2021
|
|
6WY9
 
 | | Tcur3481-Tcur3483 steroid ACAD G363A variant | | Descriptor: | Acyl-CoA dehydrogenase domain protein Tcur3481, Acyl-CoA dehydrogenase domain protein Tcur3483, CHLORIDE ION, ... | | Authors: | Kimber, M.S, Stirling, A.J, Seah, S.Y.K. | | Deposit date: | 2020-05-12 | | Release date: | 2020-10-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Key Glycine in Bacterial Steroid-Degrading Acyl-CoA Dehydrogenases Allows Flavin-Ring Repositioning and Modulates Substrate Side Chain Specificity.
Biochemistry, 59, 2020
|
|
6WY8
 
 | | Tcur3481-Tcur3483 steroid ACAD | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Acyl-CoA dehydrogenase domain protein Tcur3481, Acyl-CoA dehydrogenase domain protein Tcur3483, ... | | Authors: | Kimber, M.S, Stirling, A.J, Seah, S.Y.K. | | Deposit date: | 2020-05-12 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Key Glycine in Bacterial Steroid-Degrading Acyl-CoA Dehydrogenases Allows Flavin-Ring Repositioning and Modulates Substrate Side Chain Specificity.
Biochemistry, 59, 2020
|
|
