3L4R
 
 | | Crystal structure of the dog lipocalin allergen Can f 2 and implications for cross-reactivity to the cat allergen Fel d 4 | | Descriptor: | GLYCEROL, Minor allergen Can f 2 | | Authors: | Madhurantakam, C, Nilsson, O.B, Gronlund, H, Achour, A. | | Deposit date: | 2009-12-21 | | Release date: | 2010-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal Structure of the Dog Lipocalin Allergen Can f 2: Implications for Cross-reactivity to the Cat Allergen Fel d 4
J.Mol.Biol., 401, 2010
|
|
1U2Q
 
 | | Crystal structure of Mycobacterium tuberculosis Low Molecular Weight Protein Tyrosine Phosphatase (MPtpA) at 2.5A resolution with glycerol in the active site | | Descriptor: | CHLORIDE ION, GLYCEROL, low molecular weight protein-tyrosine-phosphatase | | Authors: | Madhurantakam, C, Rajakumara, E, Mazumdar, P.A, Saha, B, Mitra, D, Wiker, H.G, Sankaranarayanan, R, Das, A.K. | | Deposit date: | 2004-07-20 | | Release date: | 2005-03-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Low-Molecular-Weight Protein Tyrosine Phosphatase from Mycobacterium tuberculosis at 1.9-A Resolution
J.Bacteriol., 187, 2005
|
|
1U2P
 
 | | Crystal structure of Mycobacterium tuberculosis Low Molecular Protein Tyrosine Phosphatase (MPtpA) at 1.9A resolution | | Descriptor: | CHLORIDE ION, low molecular weight protein-tyrosine-phosphatase | | Authors: | Madhurantakam, C, Rajakumara, E, Mazumdar, P.A, Saha, B, Mitra, D, Wiker, H.G, Sankaranarayanan, R, Das, A.K. | | Deposit date: | 2004-07-20 | | Release date: | 2005-03-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Low-Molecular-Weight Protein Tyrosine Phosphatase from Mycobacterium tuberculosis at 1.9-A Resolution
J.Bacteriol., 187, 2005
|
|
3ROL
 
 | | Murine class I major histocompatibility complex H-2Kb in complex with post-translationally modified LCMV-derived gp34-41 peptide, comprising a nitrotyrosine at position 3 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Madhurantakam, C, Duru, A.D, Leong, C, Sandalova, T, Webb, J.R, Achour, A. | | Deposit date: | 2011-04-26 | | Release date: | 2012-04-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Nitro-tyrosination of the immunodominant LCMV epitope gp34-41 alters both its capacity to stabilize H-2Kb and the molecular surface of the MHC complex, affecting TCR recognition
PLoS ONE, 2012
|
|
3ROO
 
 | | Murine class I major histocompatibility complex H-2Kb in complex with immunodominant LCMV-derived gp34-41 peptide | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Madhurantakam, C, Duru, A.D, Leong, C, Sandalova, T, Webb, J.R, Achour, A. | | Deposit date: | 2011-04-26 | | Release date: | 2012-04-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Nitro-tyrosination of the immunodominant LCMV epitope gp34-41 alters both its capacity to stabilize H-2Kb and the molecular surface of the MHC complex, affecting TCR recognition
PLoS ONE, 2012
|
|
4DB6
 
 | | Designed Armadillo repeat protein (YIIIM3AII) | | Descriptor: | Armadillo repeat protein | | Authors: | Madhurantakam, C, Varadamsetty, G, Grutter, M.G, Pluckthun, A, Mittl, P.R.E. | | Deposit date: | 2012-01-13 | | Release date: | 2012-05-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-based optimization of designed Armadillo-repeat proteins.
Protein Sci., 21, 2012
|
|
4DB8
 
 | | Designed Armadillo-repeat Protein | | Descriptor: | Armadillo-repeat Protein, MAGNESIUM ION | | Authors: | Madhurantakam, C, Varadamsetty, G, Grutter, M.G, Pluckthun, A, Mittl, P.R.E. | | Deposit date: | 2012-01-13 | | Release date: | 2012-05-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-based optimization of designed Armadillo-repeat proteins.
Protein Sci., 21, 2012
|
|
4DBA
 
 | | Designed Armadillo repeat protein (YIIM3AII) | | Descriptor: | Designed Armadillo repeat protein, YIIM3AII, GLYCEROL | | Authors: | Madhurantakam, C, Varadamsetty, G, Grutter, M.G, Pluckthun, A, Mittl, P.R.E. | | Deposit date: | 2012-01-13 | | Release date: | 2012-05-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-based optimization of designed Armadillo-repeat proteins.
Protein Sci., 21, 2012
|
|
4DB9
 
 | | Designed Armadillo repeat protein (YIIIM3AIII) | | Descriptor: | Armadillo repeat protein, YIIIM3AIII | | Authors: | Madhurantakam, C, Varadamsetty, G, Grutter, M.G, Pluckthun, A, Mittl, P.R.E. | | Deposit date: | 2012-01-13 | | Release date: | 2012-05-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-based optimization of designed Armadillo-repeat proteins.
Protein Sci., 21, 2012
|
|
4HJ0
 
 | | Crystal structure of the human GIPr ECD in complex with Gipg013 Fab at 3-A resolution | | Descriptor: | Gastric inhibitory polypeptide receptor, Gipg013 Fab, Antagonizing antibody to the GIP Receptor, ... | | Authors: | Madhurantakam, C, Ravn, P, Gruetter, M.G, Jackson, R.H. | | Deposit date: | 2012-10-12 | | Release date: | 2013-05-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and Pharmacological Characterization of Novel Potent and Selective Monoclonal Antibody Antagonists of Glucose-dependent Insulinotropic Polypeptide Receptor.
J.Biol.Chem., 288, 2013
|
|
4V3Q
 
 | | Designed armadillo repeat protein with 4 internal repeats, 2nd generation C-cap and 3rd generation N-cap. | | Descriptor: | CALCIUM ION, GLYCEROL, YIII_M4_AII | | Authors: | Reichen, C, Madhurantakam, C, Pluckthun, A, Mittl, P. | | Deposit date: | 2014-10-20 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of designed armadillo-repeat proteins show propagation of inter-repeat interface effects.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
4V3R
 
 | | Designed armadillo repeat protein with 5 internal repeats, 2nd generation C-cap and 3rd generation N-cap. | | Descriptor: | MAGNESIUM ION, YIII_M5_AII | | Authors: | Reichen, C, Madhurantakam, C, Pluckthun, A, Mittl, P. | | Deposit date: | 2014-10-20 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of Designed Armadillo-Repeat Proteins Show Propagation of Inter-Repeat Interface Effects
Acta Crystallogr.,Sect.D, 72, 2016
|
|
4V3O
 
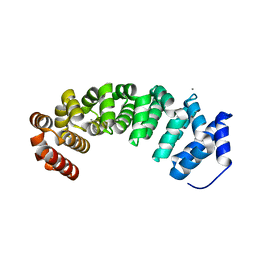 | | Designed armadillo repeat protein with 5 internal repeats, 2nd generation C-cap and 3rd generation N-cap. | | Descriptor: | ACETATE ION, CALCIUM ION, YIII_M5_AII | | Authors: | Reichen, C, Madhurantakam, C, Pluckthun, A, Mittl, P. | | Deposit date: | 2014-10-20 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of Designed Armadillo-Repeat Proteins Show Propagation of Inter-Repeat Interface Effects
Acta Crystallogr.,Sect.D, 72, 2016
|
|
5AEI
 
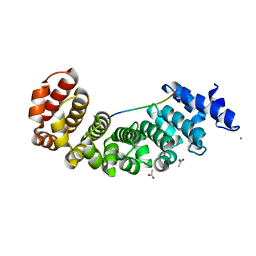 | | Designed Armadillo repeat protein YIIIM5AII in complex with peptide (KR)5 | | Descriptor: | ACETATE ION, CALCIUM ION, DESIGNED ARMADILLO REPEAT PROTEIN YIIIM5AII, ... | | Authors: | Hansen, S, Tremmel, D, Madhurantakam, C, Reichen, C, Mittl, P, Plueckthun, A. | | Deposit date: | 2015-08-31 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structure and Energetic Contributions of a Designed Modular Peptide-Binding Protein with Picomolar Affinity.
J.Am.Chem.Soc., 138, 2016
|
|
3G66
 
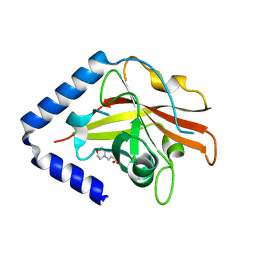 | | The crystal structure of Streptococcus pneumoniae Sortase C provides novel insights into catalysis as well as pilin substrate specificity | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Sortase C | | Authors: | Neiers, F, Madhurantakam, C, Falker, S, Manzano, C, Dessen, A, Normark, S, Henriques-Normark, B, Achour, A. | | Deposit date: | 2009-02-06 | | Release date: | 2009-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Two crystal structures of pneumococcal pilus sortase C provide novel insights into catalysis and substrate specificity.
J.Mol.Biol., 393, 2009
|
|
3G69
 
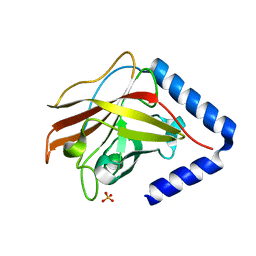 | | The crystal structure of Streptococcus pneumoniae Sortase C provides novel insights into catalysis as well as pilin substrate specificity | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, SULFATE ION, Sortase C | | Authors: | Neiers, F, Madhurantakam, C, Falker, S, Manzano, C, Dessen, A, Normark, S, Henriques-Normark, B, Achour, A. | | Deposit date: | 2009-02-06 | | Release date: | 2009-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Two crystal structures of pneumococcal pilus sortase C provide novel insights into catalysis and substrate specificity.
J.Mol.Biol., 393, 2009
|
|
4PLS
 
 | | Crystal Structures of Designed Armadillo Repeat Proteins: Implications of Construct Design and Crystallization Conditions on Overall Structure. | | Descriptor: | ACETATE ION, Arm00010, CALCIUM ION | | Authors: | Reichen, C, Madhurantakam, C, Plueckthun, A, Mittl, P.R. | | Deposit date: | 2014-05-19 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structures of Designed Armadillo Repeat Proteins: Implications of Construct Design and Crystallization Conditions on Overall Structure.
Protein Sci., 23, 2014
|
|
4PLR
 
 | | Crystal Structures of Designed Armadillo Repeat Proteins: Implications of Construct Design and Crystallization Conditions on Overall Structure. | | Descriptor: | Arm00008, CALCIUM ION | | Authors: | Reichen, C, Madhurantakam, C, Plueckthun, A, Mittl, P. | | Deposit date: | 2014-05-19 | | Release date: | 2014-08-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of designed armadillo repeat proteins: Implications of construct design and crystallization conditions on overall structure.
Protein Sci., 23, 2014
|
|
4PLQ
 
 | |
4D49
 
 | | Crystal structure of computationally designed armadillo repeat proteins for modular peptide recognition. | | Descriptor: | ARGININE, ARMADILLO REPEAT PROTEIN ARM00027, POLY ARG DECAPEPTIDE | | Authors: | Reichen, C, Forzani, C, Zhou, T, Parmeggiani, F, Fleishman, S.J, Mittl, P.R.E, Madhurantakam, C, Honegger, A, Ewald, C, Zerbe, O, Baker, D, Caflisch, A, Pluckthun, A. | | Deposit date: | 2014-10-27 | | Release date: | 2016-01-13 | | Last modified: | 2016-11-09 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Computationally Designed Armadillo Repeat Proteins for Modular Peptide Recognition.
J.Mol.Biol., 428, 2016
|
|
4D4E
 
 | | Crystal structure of computationally designed armadillo repeat proteins for modular peptide recognition. | | Descriptor: | ARMADILLO REPEAT PROTEIN ARM00016, GLYCEROL | | Authors: | Reichen, C, Forzani, C, Zhou, T, Parmeggiani, F, Fleishman, S.J, Mittl, P.R.E, Madhurantakam, C, Honegger, A, Ewald, C, Zerbe, O, Baker, D, Caflisch, A, Pluckthun, A. | | Deposit date: | 2014-10-28 | | Release date: | 2016-01-13 | | Last modified: | 2016-11-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Computationally Designed Armadillo Repeat Proteins for Modular Peptide Recognition.
J.Mol.Biol., 428, 2016
|
|
3BT4
 
 | | Crystal Structure Analysis of AmFPI-1, fungal protease inhibitor from Antheraea mylitta | | Descriptor: | Fungal protease inhibitor-1, GLYCEROL | | Authors: | Roy, S, Aravind, P, Madhurantakam, C, Ghosh, A.K, Sankarananarayanan, R, Das, A.K. | | Deposit date: | 2007-12-27 | | Release date: | 2008-12-30 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a fungal protease inhibitor from Antheraea mylitta
J.Struct.Biol., 166, 2009
|
|
3QUL
 
 | | Crystal structures of the murine class I major histocompatibility complex H-2Db in complex with LCMV-derived gp33 altered peptide ligand (Y4S) | | Descriptor: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, D-B alpha chain, ... | | Authors: | Allerbring, E, Duru, A.D, Uchtenhagen, H, Madhurantakam, C, Grimm, S, Tomek, M.B, Mazumdar, P.A, Spetz, A, Friemann, R, Sandalova, T, Uhlin, M, Nygren, P, Achour, A. | | Deposit date: | 2011-02-24 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Unexpected T-cell recognition of an altered peptide ligand is driven by reversed thermodynamics.
Eur.J.Immunol., 42, 2012
|
|
3QUK
 
 | | Crystal structures of the murine class I major histocompatibility complex H-2Db in complex with LCMV-derived gp33 altered peptide ligand (Y4A) | | Descriptor: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, D-B alpha chain, ... | | Authors: | Allerbring, E, Duru, A.D, Uchtenhagen, H, Madhurantakam, C, Grimm, S, Tomek, M.B, Mazumdar, P.A, Spetz, A, Friemann, R, Sandalova, T, Uhlin, M, Nygren, P, Achour, A. | | Deposit date: | 2011-02-24 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Unexpected T-cell recognition of an altered peptide ligand is driven by reversed thermodynamics.
Eur.J.Immunol., 42, 2012
|
|
4NSK
 
 | | CRYSTAL STRUCTURE OF THE MURINE CLASS I MAJOR HISTOCOMPATIBILITY COMPLEX H-2DB IN COMPLEX WITH LCMV-DERIVED GP33 ALTERED PEPTIDE ligand V3P | | Descriptor: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, D-B alpha chain, ... | | Authors: | Allerbring, E.B, Duru, A.D, Uchtenhagen, H, Mazumdar, P.A, Badia-Martinez, D, Madhurantakam, C, Sandalova, T, Nygren, P, Achour, A. | | Deposit date: | 2013-11-28 | | Release date: | 2014-01-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | CRYSTAL STRUCTURE OF THE MURINE CLASS I MAJOR HISTOCOMPATIBILITY COMPLEX H-2DB IN COMPLEX WITH LCMV-DERIVED GP33 ALTERED PEPTIDE ligand V3P
TO BE PUBLISHED
|
|
