1TUR
 
 | | SOLUTION STRUCTURE OF TURKEY OVOMUCOID THIRD DOMAIN AS DETERMINED FROM NUCLEAR MAGNETIC RESONANCE DATA | | Descriptor: | OVOMUCOID | | Authors: | Krezel, A.M, Darba, P, Robertson, A.D, Fejzo, J, Macura, S, Markley, J.L. | | Deposit date: | 1994-07-06 | | Release date: | 1994-10-15 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of turkey ovomucoid third domain as determined from nuclear magnetic resonance data.
J.Mol.Biol., 242, 1994
|
|
2KNE
 
 | | Calmodulin wraps around its binding domain in the plasma membrane CA2+ pump anchored by a novel 18-1 motif | | Descriptor: | ATPase, Ca++ transporting, plasma membrane 4, ... | | Authors: | Juranic, N, Atanasova, E, Filoteo, A.G, Macura, S, Prendergast, F.G, Penniston, J.T, Strehler, E.E. | | Deposit date: | 2009-08-21 | | Release date: | 2009-11-24 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Calmodulin wraps around its binding domain in the plasma membrane Ca2+ pump anchored by a novel 18-1 motif.
J.Biol.Chem., 285, 2010
|
|
3L1X
 
 | |
3L1Z
 
 | |
3L1Y
 
 | |
1YVA
 
 | | NMR solution structure of crambin in DPC micelles | | Descriptor: | Crambin | | Authors: | Ahn, H.-C, Markley, J.L. | | Deposit date: | 2005-02-15 | | Release date: | 2006-03-07 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Three-Dimensional Structure of the Water-Insoluble Protein Crambin in Dodecylphosphocholine Micelles and Its Minimal Solvent-Exposed Surface
J.Am.Chem.Soc., 128, 2006
|
|
1YV8
 
 | |
2KRE
 
 | |
1OMU
 
 | | SOLUTION STRUCTURE OF OVOMUCOID (THIRD DOMAIN) FROM DOMESTIC TURKEY (298K, PH 4.1) (NMR, 50 STRUCTURES) (REFINED MODEL USING NETWORK EDITING ANALYSIS) | | Descriptor: | OVOMUCOID (THIRD DOMAIN) | | Authors: | Hoogstraten, C.G, Choe, S, Westler, W.M, Markley, J.L. | | Deposit date: | 1995-10-11 | | Release date: | 1996-03-08 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Comparison of the accuracy of protein solution structures derived from conventional and network-edited NOESY data.
Protein Sci., 4, 1995
|
|
1OMT
 
 | | SOLUTION STRUCTURE OF OVOMUCOID (THIRD DOMAIN) FROM DOMESTIC TURKEY (298K, PH 4.1) (NMR, 50 STRUCTURES) (STANDARD NOESY ANALYSIS) | | Descriptor: | OVOMUCOID (THIRD DOMAIN) | | Authors: | Hoogstraten, C.G, Choe, S, Westler, W.M, Markley, J.L. | | Deposit date: | 1995-10-11 | | Release date: | 1996-03-08 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Comparison of the accuracy of protein solution structures derived from conventional and network-edited NOESY data.
Protein Sci., 4, 1995
|
|
2EYC
 
 | |
2EYD
 
 | |
2EYB
 
 | |
2EYA
 
 | |
2KTF
 
 | |
2L0F
 
 | |
2L0G
 
 | |
2KUH
 
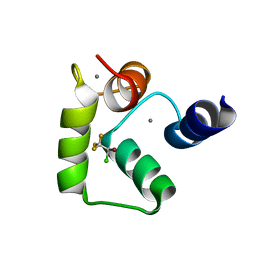 | | Halothane binds to druggable sites in calcium-calmodulin: Solution structure of halothane-CaM C-terminal domain | | Descriptor: | 2-BROMO-2-CHLORO-1,1,1-TRIFLUOROETHANE, CALCIUM ION, Calmodulin | | Authors: | Juranic, N, Macura, S, Simeonov, M.V, Jones, K.A, Penheiter, A.R, Hock, T.J, Streiff, J.H. | | Deposit date: | 2010-02-17 | | Release date: | 2010-03-09 | | Last modified: | 2013-12-11 | | Method: | SOLUTION NMR | | Cite: | Halothane binds to druggable sites in the [Ca2+]4-calmodulin (CaM) complex, but does not inhibit [Ca2+]4-CaM activation of kinase.
J. Serb. Chem. Soc., 78, 2013
|
|
2KUG
 
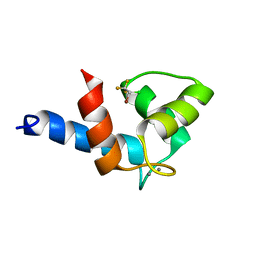 | | Halothane binds to druggable sites in calcium-calmodulin: Solution Structure of halothane-CaM N-terminal domain | | Descriptor: | 2-BROMO-2-CHLORO-1,1,1-TRIFLUOROETHANE, CALCIUM ION, Calmodulin | | Authors: | Juranic, N, Macura, S, Simeonov, M.V, Jones, K.A, Penheiter, A.R, Hock, T.J, Streiff, J.H. | | Deposit date: | 2010-02-17 | | Release date: | 2010-03-09 | | Last modified: | 2020-02-05 | | Method: | SOLUTION NMR | | Cite: | Halothane binds to druggable sites in the [Ca2+]4-calmodulin (CaM) complex, but does not inhibit [Ca2+]4-CaM activation of kinase.
J. Serb. Chem. Soc., 78, 2013
|
|
