1YIV
 
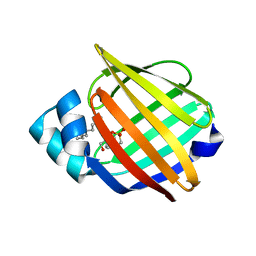 | | Structure of myelin P2 protein from Equine spinal cord | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, LAURYL DIMETHYLAMINE-N-OXIDE, Myelin P2 protein | | Authors: | Hunter, D.J.B, MacMaster, R, Rozak, A.W, Riboldi-Tunnicliffe, A, Grifiths, I.R, Freer, A.A. | | Deposit date: | 2005-01-13 | | Release date: | 2005-07-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of myelin P2 protein from equine spinal cord.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
4AQX
 
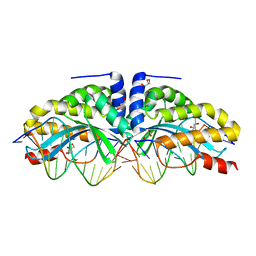 | | Crystal structure of I-CreI complexed with its target methylated at position plus 2 (in the b strand) in the presence of magnesium | | Descriptor: | 5'-D(*CP*CP*AP*AP*AP*CP*TP*GP*TP*CP*TP*CP*AP*5CM)-3', 5'-D(*GP*AP*CP*AP*GP*TP*TP*TP*GP*GP)-3', 5'-D(*GP*AP*CP*GP*TP*TP*TP*TP*GP*AP)-3', ... | | Authors: | Valton, J, Daboussi, F, Leduc, S, Redondo, P, Macmaster, R, Molina, R, Montoya, G, Duchateau, P. | | Deposit date: | 2012-04-19 | | Release date: | 2012-07-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | 5'-Cytosine-Phosphoguanine (Cpg) Methylation Impacts the Activity of Natural and Engineered Meganucleases.
J.Biol.Chem., 287, 2012
|
|
4AQU
 
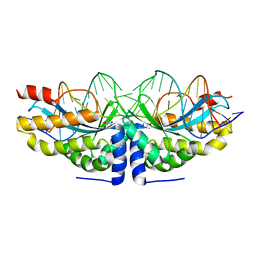 | | Crystal structure of I-CreI complexed with its target methylated at position plus 2 (in the b strand) in the presence of calcium | | Descriptor: | 5'-D(*DCP*CP*AP*AP*AP*CP*TP*GP*TP*CP*TP*CP*AP*5CMP *GP*AP*CP*GP*TP*TP*TP*TP*GP*A)-3', 5'-D(*DTP*CP*AP*AP*AP*AP*CP*GP*TP*CP*GP*TP*GP*DAP *GP*AP*CP*AP*GP*TP*TP*TP*GP*G)-3', CALCIUM ION, ... | | Authors: | Valton, J, Daboussi, F, Leduc, S, Redondo, P, Macmaster, R, Molina, R, Montoya, G, Duchateau, P. | | Deposit date: | 2012-04-19 | | Release date: | 2012-07-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | 5'-Cytosine-Phosphoguanine (Cpg) Methylation Impacts the Activity of Natural and Engineered Meganucleases.
J.Biol.Chem., 287, 2012
|
|
3MQ2
 
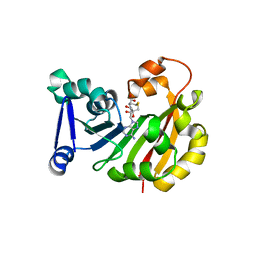 | | Crystal Structure of 16S rRNA Methyltranferase KamB | | Descriptor: | 16S rRNA methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Macmaster, R.A. | | Deposit date: | 2010-04-27 | | Release date: | 2010-12-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural insights into the function of aminoglycoside-resistance A1408 16S rRNA methyltransferases from antibiotic-producing and human pathogenic bacteria.
Nucleic Acids Res., 38, 2010
|
|
3MTE
 
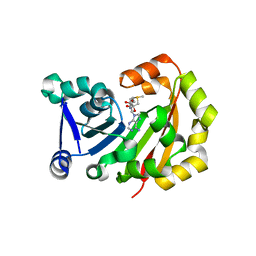 | | Crystal Structure of 16S rRNA Methyltranferase | | Descriptor: | 16S rRNA methylase, S-ADENOSYLMETHIONINE | | Authors: | Macmaster, R.A, Conn, G.L, Zelinskaya, N. | | Deposit date: | 2010-04-30 | | Release date: | 2010-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.805 Å) | | Cite: | Structural insights into the function of aminoglycoside-resistance A1408 16S rRNA methyltransferases from antibiotic-producing and human pathogenic bacteria.
Nucleic Acids Res., 38, 2010
|
|
2H8C
 
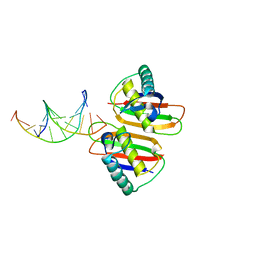 | | Structure of RusA D70N in complex with DNA | | Descriptor: | 5'-D(*CP*CP*GP*GP*TP*AP*CP*CP*GP*GP*T)-3', Crossover junction endodeoxyribonuclease rusA | | Authors: | Macmaster, R.A. | | Deposit date: | 2006-06-07 | | Release date: | 2007-04-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | RusA Holliday junction resolvase: DNA complex structure--insights into selectivity and specificity.
Nucleic Acids Res., 34, 2006
|
|
2H8E
 
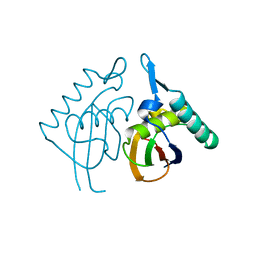 | | Structure RusA D70N | | Descriptor: | Crossover junction endodeoxyribonuclease rusA | | Authors: | Macmaster, R.A. | | Deposit date: | 2006-06-07 | | Release date: | 2007-04-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | RusA Holliday junction resolvase: DNA complex structure--insights into selectivity and specificity.
Nucleic Acids Res., 34, 2006
|
|
4RWZ
 
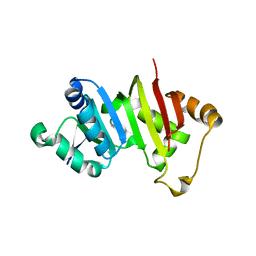 | | Crystal structure of the antibiotic-resistance methyltransferase Kmr | | Descriptor: | Putative rRNA methyltransferase | | Authors: | Savic, M. | | Deposit date: | 2014-12-08 | | Release date: | 2015-03-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | 30S Subunit-Dependent Activation of the Sorangium cellulosum So ce56 Aminoglycoside Resistance-Conferring 16S rRNA Methyltransferase Kmr.
Antimicrob.Agents Chemother., 59, 2015
|
|
4RX1
 
 | | Crystal Structure of antibiotic-resistance methyltransferase Kmr | | Descriptor: | GLYCEROL, IODIDE ION, Putative rRNA methyltransferase | | Authors: | Savic, M. | | Deposit date: | 2014-12-08 | | Release date: | 2015-03-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.472 Å) | | Cite: | 30S Subunit-Dependent Activation of the Sorangium cellulosum So ce56 Aminoglycoside Resistance-Conferring 16S rRNA Methyltransferase Kmr.
Antimicrob.Agents Chemother., 59, 2015
|
|
