1JAJ
 
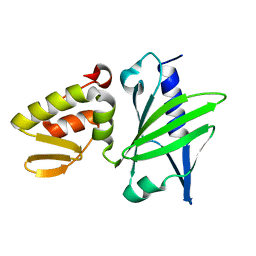 | |
1DK3
 
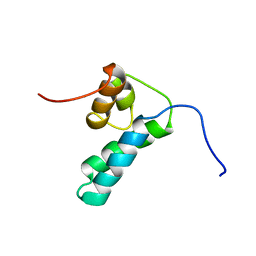 | | REFINED SOLUTION STRUCTURE OF THE N-TERMINAL DOMAIN OF DNA POLYMERASE BETA | | Descriptor: | DNA POLYMERASE BETA | | Authors: | Maciejewski, M.W, Prasad, R, Liu, D.-J, Wilson, S.H, Mullen, G.P. | | Deposit date: | 1999-12-06 | | Release date: | 2000-02-14 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Backbone dynamics and refined solution structure of the N-terminal domain of DNA polymerase beta. Correlation with DNA binding and dRP lyase activity.
J.Mol.Biol., 296, 2000
|
|
1DK2
 
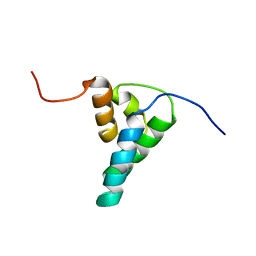 | | REFINED SOLUTION STRUCTURE OF THE N-TERMINAL DOMAIN OF DNA POLYMERASE BETA | | Descriptor: | DNA POLYMERASE BETA | | Authors: | Maciejewski, M.W, Prasad, R, Liu, D.-J, Wilson, S.H, Mullen, G.P. | | Deposit date: | 1999-12-06 | | Release date: | 2000-02-14 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Backbone dynamics and refined solution structure of the N-terminal domain of DNA polymerase beta. Correlation with DNA binding and dRP lyase activity.
J.Mol.Biol., 296, 2000
|
|
1M9L
 
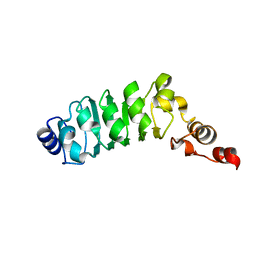 | | Relaxation-based Refined Structure Of Chlamydomonas Outer Arm Dynein Light Chain 1 | | Descriptor: | Outer Arm Dynein Light Chain 1 | | Authors: | Wu, H.W, Maciejewski, M.W, Marintchev, A, Benashski, S.E, Mullen, G.P, King, S.M. | | Deposit date: | 2002-07-29 | | Release date: | 2003-03-04 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Relaxation-based structure refinement and backbone molecular dynamics of the
Dynein motor domain-associated light chain
Biochemistry, 42, 2003
|
|
1TVJ
 
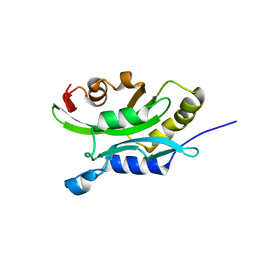 | | Solution Structure of chick cofilin | | Descriptor: | Cofilin | | Authors: | Gorbatyuk, V.Y, Nosworthy, N.J, Robson, S.A, Maciejewski, M.W, dos Remedios, C.G, King, G.F. | | Deposit date: | 2004-06-29 | | Release date: | 2005-09-20 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | NMR Study of the Molecular Basis for Phosphoinositide Regulation of the Cofilin-Actin Interactions.
To be Published
|
|
2H1Z
 
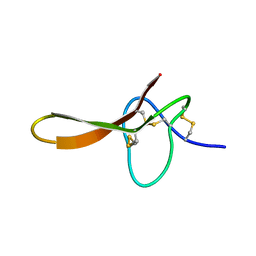 | |
1DS9
 
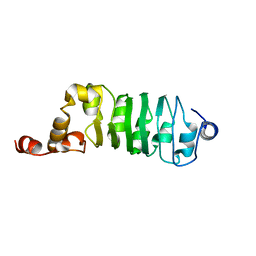 | | SOLUTION STRUCTURE OF CHLAMYDOMONAS OUTER ARM DYNEIN LIGHT CHAIN 1 | | Descriptor: | OUTER ARM DYNEIN | | Authors: | Wu, H.W, Maciejewski, M.W, Marintchev, A, Benashski, S.E, Mullen, G.P, King, S.M. | | Deposit date: | 2000-01-07 | | Release date: | 2000-07-26 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a dynein motor domain associated light chain.
Nat.Struct.Biol., 7, 2000
|
|
1U3O
 
 | | Solution structure of rat Kalirin N-terminal SH3 domain | | Descriptor: | Huntingtin-associated protein-interacting protein | | Authors: | Schiller, M.R, Chakrabarti, K, King, G.F, Schiller, N.I, Eipper, B.A, Maciejewski, M.W. | | Deposit date: | 2004-07-22 | | Release date: | 2005-07-26 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Regulation of RhoGEF Activity by Intramolecular and Intermolecular SH3 Domain Interactions.
J.Biol.Chem., 281, 2006
|
|
1XDX
 
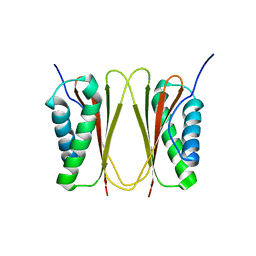 | |
2N67
 
 | |
2M2I
 
 | |
2M19
 
 | | Solution structure of the Haloferax volcanii HVO 2177 protein | | Descriptor: | Molybdopterin converting factor subunit 1 | | Authors: | Li, Y, Maciejewski, M.W, Martin, J, Jin, K, Zhang, Y, Lu, M, Maupin-Furlow, J.A, Hao, B. | | Deposit date: | 2012-11-21 | | Release date: | 2013-08-07 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Crystal structure of the ubiquitin-like small archaeal modifier protein 2 from Haloferax volcanii.
Protein Sci., 22, 2013
|
|
1PV0
 
 | | Structure of the Sda antikinase | | Descriptor: | Sda | | Authors: | Rowland, S.L, Burkholder, W.F, Maciejewski, M.W, Grossman, A.D, King, G.F. | | Deposit date: | 2003-06-26 | | Release date: | 2004-04-13 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Structure and mechanism of Sda: an inhibitor of the histidine kinases that regulate initiation of sporulation in Bacillus subtilis
Mol.Cell, 13, 2004
|
|
1PZ7
 
 | | Modulation of agrin function by alternative splicing and Ca2+ binding | | Descriptor: | Agrin, CALCIUM ION | | Authors: | Stetefeld, J, Alexandrescu, A.T, Maciejewski, M.W, Jenny, M, Rathgeb-Szabo, K, Schulthess, T, Landwehr, R, Frank, S, Ruegg, M.A, Kammerer, R.A. | | Deposit date: | 2003-07-10 | | Release date: | 2004-04-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.421 Å) | | Cite: | Modulation of agrin function by alternative splicing and Ca2+ binding.
STRUCTURE, 12, 2004
|
|
1PZ9
 
 | | Modulation of agrin function by alternative splicing and Ca2+ binding | | Descriptor: | Agrin | | Authors: | Stetefeld, J, Alexandrescu, A.T, Maciejewski, M.W, Jenny, M, Rathgeb-Szabo, K, Schulthess, T, Landwehr, R, Frank, S, Ruegg, M.A, Kammerer, R.A. | | Deposit date: | 2003-07-10 | | Release date: | 2004-04-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Modulation of agrin function by alternative splicing and Ca2+ binding.
STRUCTURE, 12, 2004
|
|
1Q56
 
 | | NMR structure of the B0 isoform of the agrin G3 domain in its Ca2+ bound state | | Descriptor: | Agrin | | Authors: | Stetefeld, J, Alexandrescu, A.T, Maciejewski, M.W, Jenny, M, Rathgeb-Szabo, K, Schulthess, T, Landwehr, R, Frank, S, Ruegg, M.A, Kammerer, R.A. | | Deposit date: | 2003-08-06 | | Release date: | 2004-04-13 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Modulation of agrin function by alternative splicing and Ca2+ binding
Structure, 12, 2004
|
|
1PZ8
 
 | | Modulation of agrin function by alternative splicing and Ca2+ binding | | Descriptor: | Agrin, CALCIUM ION | | Authors: | Stetefeld, J, Alexandrescu, A.T, Maciejewski, M.W, Jenny, M, Rathgeb-Szabo, K, Schulthess, T, Landwehr, R, Frank, S, Ruegg, M.A, Kammerer, R.A. | | Deposit date: | 2003-07-10 | | Release date: | 2004-04-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Modulation of agrin function by alternative splicing and Ca2+ binding.
STRUCTURE, 12, 2004
|
|
1EV0
 
 | |
6O59
 
 | |
5IIR
 
 | |
5IIV
 
 | | GCN4p pH 1.5 | | Descriptor: | General control protein GCN4 | | Authors: | Brady, M.R, Kaplan, A.R, Alexandrescu, A.T. | | Deposit date: | 2016-03-01 | | Release date: | 2017-03-15 | | Last modified: | 2017-04-05 | | Method: | SOLUTION NMR | | Cite: | Nuclear Magnetic Resonance Structures of GCN4p Are Largely Conserved When Ion Pairs Are Disrupted at Acidic pH but Show a Relaxation of the Coiled Coil Superhelix.
Biochemistry, 56, 2017
|
|
5IEW
 
 | |
1XNA
 
 | |
1XNT
 
 | |
4HRO
 
 | |
